The Cancer Genome Atlas output and practice of gastric cancer
Any clinical professionals who devote themselves to prevention, diagnosis, therapy, and management of gastric cancer patients are now again facing another achievement by the consortium of The Cancer Genome Atlas (TCGA) (1). In the era of post human genome sequence and massive parallel sequencing technology, every week this kind of huge data draw a transient attention of our colleagues who are very busy with conventional routines. Information like this on the cutting edge of science is not usually related to the action plan next week at the clinic. In a field of lung cancer managements, however, we witnessed the latest fruits of these technologies such as series of discoveries of targetable fused-kinase protein drastically changed clinical practice (2). How influential the results presented here in this article can be to the practice today, tomorrow, and in future? Reasonably, clinicians in any fields of specific organ cancers hope categorization of cancers based on the state-of-the-art technology can specify the fittest therapy in each individual. In the field of gastric cancer research, attempts to delineate genomic characteristics of gastric cancer and to take advantage of these features as potential targets of therapy have been popular in the literatures of the last few years.
For example, Kubo et al. reported re-sequencing and copy number analysis of kinases in gastric cancer (3) and Kiyose et al. further applied 400 BAC FISH probes on the tissue microarray of 350 gastric cancers, identified several kinase gene amplification, and suggested the assays could be used as companion diagnosis on pathology archives like Hercep TestTM (4). Hillmer et al. applied paired-end-tag sequencing approach to four gastric cancers and found structural variations in them (5). Zang et al. focused on kinase changes in 14 gastric cancer cell lines (6). Methodologies were various. Deng et al. investigated 193 primary gastric tumors by high resolution SNP array and copy number changes in the tumors. Based on the huge mutational information of gastric cancer obtained by massively parallel short read and DNA paired-end tag sequencing, Nagarajan et al. tried to classify gastric cancers into two categories; microsatellite instability-positive gastric cancer and TP53-wild type cancer (7). Then Zang et al. did exome analysis of 15 cases and disclosed mutations of chromatin modifier genes such as ARID1A and cell adhesion molecule such as FAT4 (8). As to the MSI positive fraction of gastric cancer, Korean researchers extensively clarified mutation profile (9). In the course of rapid popularity of “genome-wide” approaches applied to each cancer case, a peculiar pathological status became clarified such as GLO amplification as a new metabolic marker of gastric cancer (10).
In the paper published in September issue of Nature, TCGA reported the landscape of somatic changes in gastric cancer in comprehensive way. The data they showed include mutations per Mb, copy number changes (somatic copy number alteration SCNA), DNA methylation, mRNA expression profile, micro RNA profile analysis, microsatellite instability, Epstein-Bar virus infection status as well as whole genome analysis for identification of structural changes (such as fusion genes) found in gastric cancer. According to the supplementary table of this paper, out of 295 cases, the cases with T1A and T1B are 11 (3.7%) and the T3 cases are about half of the total cases. This fact implies the idea and consequent strategy for gastric cancer therapy generated by this study are mainly applicable to T3 and T4, an advanced stage gastric cancer, some of which are inoperable. For example, the managements widely recommended in Japan, that is, detection of gastric cancer at early stage by intensive surveillance and endoscopical submucosal dissections (Figure 1) for nearly asymptomatic subjects (covered by government-based health insurance), are out of the scope of this costly analysis and therapeutic plan based on it they envisaged here.
The tremendous data set published by TCGA suggested four categories of gastric cancer: (I) EBV positive cases; (II) microsatellite instability (MSI) positive cases; (III) chromosomal instability (CIN) type; (IV) genomically-stable type (near-diploid type). The hallmarks of these four groups can be said in another way: (I) hypermethylation type; (II) KRAS, PTEN, PIK3CA mutations; (III) kinase receptor amplifications; (IV) diffuse type with RHOA mutation, respectively.
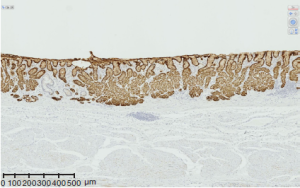
For the last two decades, the above four aspects of gastric cancer have been repeatedly investigated both sporadically and systematically in various scale of projects including the very recent studies by two group which identified RHOA mutation (11,12). The genes where mutations were more frequently found than RHOA are TP53, CDH1, SMAD4, and PIK3CA which are consistent with the previous reports, and ARID1A, KRAS, MUC6, and APC followed. The authors highlighted the PI3KCA mutations, extreme methylation, and amplifications of JAK2, PD-L1, and PDL2 in EBV positive category. As for fusion genes, transcripts involving CLDN18i, which is specifically expressed in gastric epithelium (Figure 2) were detected. Its partners were ARHGAP family genes. The genes involved in this and related pathways have been investigated for years such as involvement of ARHGEF 6 (beta-PIX) and ARHGEF (alpha-PIX) by the researchers of cell signaling (13-16) and the involvement of ARHGAP 6 and 26 in this TCGA paper are mechanistically understandable, especially considering these were found in diffuse type, notoriously invasive subtype of gastric cancers. The finding that the 5’ side of fusion transcript is CLDN18, a claudin specifically expressed in the stomach reminds us that SLC34A2 specifically expressed in type II alveolar cell of the lung has been found as a component of fusion transcript in some of lung cancers (17,18).
Based on these data, the authors encourage the readers, and probably themselves, by pointing out that the signaling molecules above-mentioned could be targetable. The involvement of PD-1 and 2, immune checkpoint inhibitors, in EBV related gastric cancer is remarkable considering these molecules are enthusiastically promoted as targets of immunotherapy (especially in malignant melanoma) (19). Obviously the practical feasibility of the management of gastric cancer based on the proposals of this paper warrants further applied and translational researches and assessments by several sectors including academics, industries, health insurance companies, and attending doctors.
The other point to bewilder the practical pathologists is histological sub-classification shown in the supplementary table (1). Sub-classification of gastric cancer ranged from Lauren dichotomy (actually this paper adopts a trichotomy including mixed type) to the Japanese classification systems (http://www.jgca.jp/pdf/JGCA_Jpn_Classification_3rd_Eng.pdf, 2011) which morphologically scrutinize very minute attributes up to the level where it sometimes suffers from the Galapagos Syndrome—it has evolved separately from the rest of the world. WHO system would be a wise and modest way when describing the statistics. The most pathologists, however, are very familiar with the morphological heterogeneity in single tumor in advanced stage gastric cancer especially where several blocks (five and more, sometimes 30 to 50) are routinely made for pathological investigation. As expected, the histological sub-classification itself was not related to molecular signature shown here. Thus the cancer, a real challenge we should treat may evade “individualized” therapeutic strategy this ambitious presentation proposes. On the other hand, in the next stage, the application of the tour de force genetic analyses to the initial stage of gastric carcinogenesis will further provide efficient predictive and preventive measures of this ominous cancer.
Acknowledgements
We greatly appreciate the grants from the Ministry of Health, Labour and Welfare (21-1,10103838), from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (221S0001), from Princess Takamatsu Cancer Research Fund, and from the Smoking Research Foundation.
Disclosure: The author declares no conflict of interest.
References
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [PubMed]
- Kubo T, Kuroda Y, Shimizu H, et al. Resequencing and copy number analysis of the human tyrosine kinase gene family in poorly differentiated gastric cancer. Carcinogenesis 2009;30:1857-64. [PubMed]
- Kiyose S, Nagura K, Tao H, et al. Detection of kinase amplifications in gastric cancer archives using fluorescence in situ hybridization. Pathol Int 2012;62:477-84. [PubMed]
- Hillmer AM, Yao F, Inaki K, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res 2011;21:665-75. [PubMed]
- Zang ZJ, Ong CK, Cutcutache I, et al. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res 2011;71:29-39. [PubMed]
- Nagarajan N, Bertrand D, Hillmer AM, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol 2012;13:R115. [PubMed]
- Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 2012;44:570-4. [PubMed]
- Yoon K, Lee S, Han TS, et al. Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers. Genome Res 2013;23:1109-17. [PubMed]
- Hosoda F, Arai Y, Okada N, et al. Integrated genomic and functional analyses reveal glyoxalase I as a novel metabolic oncogene in human gastric cancer. Oncogene 2014. [Epub ahead of print]. [PubMed]
- Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573-82. [PubMed]
- Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583-7. [PubMed]
- Zy Li, Yj Wang, Jp Song, et al. Genomic structure of the human beta-PIX gene and its alteration in gastric cancer. Cancer Lett 2002;177:203-8. [PubMed]
- Yoshii S, Tanaka M, Otsuki Y, et al. Involvement of alpha-PAK-interacting exchange factor in the PAK1-c-Jun NH(2)-terminal kinase 1 activation and apoptosis induced by benzo[a]pyrene. Mol Cell Biol 2001;21:6796-807. [PubMed]
- Wang YJ, Oba SM, Yoshii S, et al. Genomic structure of human alpha-pix, and variable deletions in a poly (T) tract in gastric cancer tissue. Cancer Lett 2001;164:69-75. [PubMed]
- Yoshii S, Tanaka M, Otsuki Y, et al. alphaPIX nucleotide exchange factor is activated by interaction with phosphatidylinositol 3-kinase. Oncogene 1999;18:5680-90. [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [PubMed]
- Hashimoto M, Wang DY, Kamo T, et al. Isolation and localization of type IIb Na/Pi cotransporter in the developing rat lung. Am J Pathol 2000;157:21-7. [PubMed]
- Dolan DE, Gupta S. PD-1 pathway inhibitors: changing the landscape of cancer immunotherapy. Cancer Control 2014;21:231-7. [PubMed]


