Tumour suppressor genes in gastric carcinogenesis: an up-to-date review of genetic and epigenetic changes in the development of gastric adenocarcinoma
Introduction
Gastric cancer is the fifth commonest cancer worldwide, with approximately 1 million new cases diagnosed in 2012 (1). More than 70% of cases occur in developing countries, mainly in Eastern Asia (1). It is the third leading cause of cancer-related death, with a 5-year survival of just 20%, which is partly attributable to our relatively poor understanding of the pathogenesis of gastric cancer (1-3). The commonest type of gastric cancer is adenocarcinoma, which accounts for over 95% of diagnoses (4).
Gastric adenocarcinoma is a heterogeneous, multifactorial disease (5). Risk factors include male gender, increasing age, a diet high in salty food, pickled food, red meat or nitrosamines, infection with Helicobacter pylori (H. pylori), smoking, previous gastric ulcer or gastric polyps, pernicious anaemia, previous vagotomy or partial gastrectomy, family history, history of other cancers, radiation exposure, immunosuppression, occupational exposure and blood group A (4,6). Of these, H. pylori infection, particularly cytotoxin-associated gene A-positive [CagA (+)] H. pylori, is the most important risk factor for gastric cancer and is responsible for at least 95% of cases (7,8).
The Lauren classification divides gastric adenocarcinoma into two main histological types: intestinal or diffuse (9,10). The pathophysiology of intestinal type gastric cancer is relatively well understood and involves the transition through chronic gastritis, atrophy, intestinal metaplasia, dysplasia, intramucosal carcinoma to invasive cancer (9). By comparison, the development of diffuse gastric cancer is less well understood (9). Solcia et al. suggested that diffuse cancer develops from hyperplastic, non-metaplastic gastric glands and involves genes involved in cell-cell and cell-matrix interactions (11). A study by Nakayama et al. found that there were frequent differences in the genetic lineages between early [invasive gastric cancer that invades no more deeply than the submucosa, irrespective of lymph node metastases (T1, any N)] and advanced tubular gastric adenocarcinomas suggesting that some early cancers may not necessarily develop into advanced cancers and may instead represent a distinct variant of cancer (12).
Tumour suppressor genes (TSGs) in cancer
TSGs are genes that normally play a protective role in preventing the malignant transformation of cells by repairing DNA, inhibiting cell proliferation and initiating programmed cell death (apoptosis). They are involved in the regulation of a range of cell functions including cell adhesion, cell-cell interaction, cytoplasmic signal transduction and nuclear transcription (13).
The first identified TSG was retinoblastoma 1 (RB1) gene, which was discovered by Cavenee et al. in 1983 (14). This confirmed the “two-hit” hypothesis proposed by Knudson a decade earlier, based on observation of retinoblastoma cases and published reports, that retinoblastoma arises as a result of two mutational events (15). Several authors have since established that RB1 is lost or inactivated in many other more common human cancers including bladder, breast and lung carcinoma and that it is also a target for oncogenic proteins of DNA tumour viruses which may bind to RB family proteins inhibiting their activity at protein, rather than at gene, level (16,17).
Over recent decades, there has been a rapid expansion in the number of TSGs that have been identified in relation to a broad range of inherited and non-inherited human cancers and it has been suggested that alterations in TSGs account for some of the most common molecular changes in human carcinogenesis (18). It is hoped that greater understanding of the pattern of TSG expression in gastric cancer may enable the identification of specific biomarkers that could be used for early diagnosis and the development of targeted treatments.
Mutations in TSGs may result in their loss or inactivation, enabling cells to undergo uncontrolled cell division or malignant change resulting in the development of many types of human cancer. Most TSG mutations are acquired, although they may be inherited.
TSGs in gastric cancer
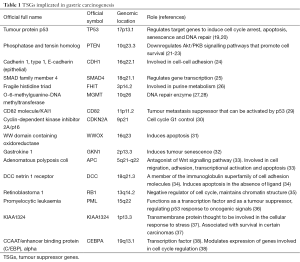
A number of TSGs have been implicated in gastric carcinogenesis (see Table 1).
Full table
A study by Lee et al. found that TSGs accounted for the majority of proteins significantly associated with survival in gastric carcinomas (39). Specifically, the overexpression of p53 and loss of expression of PTEN, E-cadherin, smad4, FHIT, MGMT and CD82 were all significantly associated with poor prognosis in gastric carcinomas (39). Loss of smad4 expression was more common in intestinal type, whilst loss of FHIT and E-cadherin expression was more common in diffuse type gastric carcinoma (39). Loss of p53 expression was more common in poorly differentiated tumours (39). Loss of E-cadherin, smad4, CD82, MGMT and PTEN expression was associated with advanced stage of disease whilst loss of FHIT and p16 expression was seen in both early and advanced stages of disease (39). Moreover, loss of TSG expression accumulates in a stepwise fashion during gastric cancer tumour progression and there is a significant correlation between this accumulation and patient survival (39). Lee et al. suggest that TSG expression status is one of the most important factors in determining the prognosis of gastric cancer (39).
In addition to mutations in TSG, genetic instability, activation of oncogenes and aberrant growth factor expression/receptor activation all play a role in gastric carcinogenesis (40). For example, in Epstein-Barr virus-associated gastric carcinoma (EBVaGC), which accounts for 10% of all gastric carcinomas, global CpG island hypermethylation occurs via viral proteins or microRNAs (miRNAs) resulting in epigenetic silencing of TSGs (41,42).
Here we discuss some of the main TSGs that have been implicated in the development of gastric adenocarcinoma and present an up-to-date review of what is currently understood about the genetic and epigenetic mediated changes that may underpin gastric carcinogenesis.
TP53
Abnormalities in TP53 have been found in over half of human cancers including leukaemia, breast, colon and lung carcinoma, and TP53 mutations are the most common genetic alteration seen in human cancer (18). They are identified in over 60% of human gastric carcinomas, regardless of histological type (43,44). The p53 protein plays a key role in cell cycle progression, preventing G1/S phase transition after DNA damage has occurred, allowing DNA repair or cell apoptosis (19). Downstream targets of p53 have also been shown to play a role in gastric cancer invasion (45).
Some studies have shown that abnormalities in TP53 can occur in non-neoplastic gastric mucosa with intestinal metaplasia suggesting that TP53 mutations may occur early in gastric carcinogenesis, whilst others have suggested that TP53 gene mutations occur relatively late in the sequence of events underlying the development of gastric carcinoma (46,47). There is some variation depending on the histological type of gastric cancer, with TP53 mutations seen early in the development of intestinal type gastric cancer but late in the development of diffuse gastric cancer (11,39).
Hamada et al. analyzed the surgical specimens of patients with gastric cancer and found that there were significantly fewer apoptotic cells following pre-operative chemotherapy or radiotherapy in tumours with mutant TP53 expression compared to those expressing wild-type TP53, suggesting that TP53 mutations are associated with poorer gastric cancer response to chemotherapy and radiotherapy (48). Moreover, mutant TP53 is associated with shorter life expectancy, not only in gastric cancer, but also in breast, lung and colorectal cancer (19).
Recent studies have suggested that H. pylori CagA (+) contributes to the inactivation of TP53 (49). The CagA protein is encoded by the cag pathogenicity island of H. pylori and interferes with the p53 tumour suppressor pathway resulting in an anti-apoptotic effect (50). CagA also targets the tumour suppressor ASPP2 (50).
PTEN
PTEN is a lipid phosphatase that dephosphorylates phosphatidylinositol-3,4,5-triphosphate (PIP3), reducing the activation of phosphoinositide 3-kinase (PI3K) and the serine/threonine-specific protein kinase Akt, both of which promote cell survival (21). Loss of PTEN increases levels of PIP3, increases Akt activation and consequently inhibits apoptosis (22).
Inactivation of PTEN may occur as a result of gene mutation, loss of heterozygosity (LOH), promoter hypermethylation or miRNA-mediated alteration in gene expression or post-translational modification and has been shown to occur in many cases of gastric cancer (51).
In gastric adenocarcinoma, mutation of PTEN is associated with more advanced stage of disease, poorer tumour differentiation, lymphatic vessel invasion and tumour chemoresistance (39,51,52). Wen et al. found that PTEN and E-cadherin protein expression are significantly downregulated in gastric cancer tissue compared with normal gastric tissue whilst PI3K, Akt, MMP-2, MMP-9 and NF-κBp65 protein are overexpressed in gastric cancer (52).
CDH1
E-cadherin has been implicated in early gastric carcinogenesis, tumour progression, invasion and metastasis (7,53). Deregulation of E-cadherin may occur as a consequence of CDH1 gene mutation, epigenetic promoter hypermethylation, LOH, transcriptional modification and regulation by miRNAs (53). Somatic alterations in CDH1 are found in approximately 30% of all patients with gastric cancer of both histological types with approximately one third of these having structural alterations (7.5% LOH, 1.7% mutation) with the remainder having epigenetic modifications (18.4% hypermethylation) (54). Epigenetic modifications are more common than structural in diffuse gastric cancer whilst intestinal type cancer has roughly similar rates of structural and epigenetic changes (54). Structural alterations, rather than epigenetic modifications, of CDH1 are associated with the poorest prognosis (54).
Loss of E-cadherin expression resulting from CDH1 gene alteration is associated with increased cancer cell proliferation, invasion and/or metastasis and is thought to be the primary carcinogenic event in hereditary diffuse gastric cancer (53,55,56). The cumulative lifetime risk of developing gastric cancer in CDH1 mutation carriers is quoted by Chun and Ford [2012] as being up to 80% (3). The level of CDH1 hypermethylation is also higher in gastric cancer and adjacent gastric mucosa than in normal gastric tissue (55). Furthermore, hypermethylation of CDH1 correlates with H. pylori status in gastric cancer (55). E-cadherin interacts with β-catenin, a proto-oncogene, at the cell-membrane and aberrant expression of this complex is also seen in gastric cancer (7).
A recent study by Wei et al. found that mRNA and protein expression levels of T-cadherin (another member of the cadherin family) are significantly lower in gastric cancer tissue compared with corresponding adjacent normal gastric tissue (57). Moreover, decreased T-cadherin protein expression is correlated with larger tumour size, lymph node metastases and higher TNM stage (57). Multivariate analysis reveals that T-cadherin expression is an independent prognostic factor for overall survival (57).
SMAD4
Mutations in SMAD4 are associated with gastric cancer (39,58). Loss of expression of smad4 is associated with advanced pT stage and poorer outcome in gastric carcinoma patients and is more common in intestinal compared with diffuse type gastric adenocarcinomas (39).
FHIT/FRA3B locus
The FHIT gene is located at the most fragile human chromosome site, a location which predisposes genes to chromosomal rearrangements that may result in neoplastic growth (39,59). LOH for alleles in this region is seen in around 50% of uncultured stomach and colon carcinomas and it is also lost or absent in lung, kidney and cervical carcinomas (59,60). Loss of FHIT protein expression is a significant and independent predictor of survival in gastric adenocarcinoma and is significantly correlated with diffuse type, poor differentiation and advanced stage of disease (39,61).
MGMT
MGMT plays an important role in the repair of DNA damage and inactivation of MGMT by promoter methylation is implicated in gastric carcinogenesis (62,63). A study of gastric cancer patients by Park et al. found that MGMT promoter methylation was significantly associated with KRAS mutation, lymph node invasion, tumour stage and disease free survival (64). The MGMT promoter is more frequently methylated in advanced gastric cancer but has also been found in early gastric cancer and precancerous lesions (65,66).
CD82 molecule (CD82)
The CD82 molecule is a membrane glycoprotein that belongs to the transmembrane 4 superfamily, which act as inhibitors of tumour cell motility and metastasis (67). Reduced expression of CD82 is found in gastric cancer and loss of CD82 protein expression is associated with disease progression, metastasis and poor clinical outcome (68,69).
CDKN2A/p16
CDKN2A is more commonly methylated in gastric cancer compared to normal gastric tissue (70).
WW domain containing oxidoreductase (WWOX)
WWOX is a TSG, involved in the negative regulation of cell growth, which is frequently mutated in gastric cancer (71,72). It is thought to play a role in tumour progression by regulating the TGFβ/SMAD and Wnt signaling pathways and loss of WWOX expression is associated with more aggressive disease and poorer prognosis in a range of cancers including gastric carcinoma (71,73,74). Increased hypermethylation of WWOX is found in EBVaGC and in the presence of H. pylori infection (75,76).
Gastrokine 1 (GKN1)
GKN1 is a TSG, highly expressed in normal stomach (77). GKN1 expression is frequently lost in the presence of H. pylori infection and gastric cancer, particularly the diffuse subtype, with conflicting evidence on its expression in metaplasia (77-79). Reduced or lost GKN1 expression occurs with similar frequency in gastric adenomas and carcinomas, suggesting it may play a role in the early stages of gastric carcinogenesis (80). GKN1 plays a role in the inflammatory response of gastric mucosa via the regulation of cytokine production, the NF-κB signaling pathway and cyclooxygenase-2 expression, inhibits the carcinogenic effect of H. pylori CagA (+) and has a pro-antioxidant effect (78). GKN1 negatively regulates cell survival, proliferation, colony formation, epithelial-to-mesenchymal transition (EMT), cancer cell migration, invasion and metastases, and promotes apoptotic cell death (78,81). Moreover, expression of GKN1 is associated with increased tumour sensitivity to fluorouracil treatment (82). Loss of GKN1 is associated with significantly shorter survival in intestinal type gastric cancer (79).
Adenomatous polyposis coli (APC)
The APC gene is located on the long arm of chromosome 5 close to other TSGs involved in gastric adenocarcinoma (83). Inactivation of APC as a consequence of germline mutations is seen in familial adenomatous polyposis (FAP) and as a result of somatic mutations in the development of a range of human cancers (84). Unlike colorectal cancer, changes in genes such as APC, though present in gastric carcinoma, are rare in gastric adenomas, suggesting that the adenoma-carcinoma sequence is less common in gastric carcinogenesis and indeed Tahara [2004] suggest that the sequence is found in only 20% of gastric adenomas with APC mutations (85,86). APC can also be inactivated by promoter hypermethylation (87).
Allele loss on chromosome 5q is often seen in gastric adenocarcinoma, particularly in well-differentiated early cancer (44). LOH at the APC genetic locus is found in over 25% of cases of gastric cancer, including early cancers, suggesting that abnormalities in the APC gene may occur early in tumourigenesis (88). In advanced gastric cancer, LOH at the APC locus is found in 30% of informative cases (89).
DCC netrin 1 receptor (DCC)
LOH at the DCC locus is associated with well differentiated and advanced gastric cancer (89,90).
RB1
RB1 is involved in the negative regulation of the cell cycle at the G1/S transition (16,91). LOH at the RB1 locus is found in 30% of informative cases of advanced gastric cancer (89). RB1 has also been shown to be a target of miRNA-106b~25, which promotes cell proliferation, EMT, cell-cycle progression and exerts an anti-apoptotic effect (92).
Promyelocytic leukaemia (PML)
PML protein is a tumour suppressor, involved in the control of apoptosis and cell proliferation via p73 and the yes-associated protein (YAP 1), which is downregulated in gastric cancer (93).
KIAA1324
KIAA1324 is downregulated in gastric cancer and plays a role in the control of cell proliferation, invasion and apoptosis as well as inhibiting GRP78 oncoprotein (94).
CCAAT/enhancer binding protein (C/EBP), alpha (CEBPA)
CEBPA encodes a transcription factor involved in inducing terminal differentiation and in the control of cell proliferation (95). It has been identified as a TSG in several types of cancer and its expression is lost in 30% of cases of gastric cancer (95).
Candidate TSGs
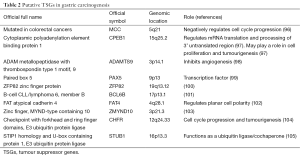
Over recent years, several putative TSG have been identified. These are shown in Table 2 and discussed below.
Full table
Mutated in colorectal cancers (MCC)
The MCC gene, like APC, is mapped to chromosome 5. A study of 24 surgical specimens of primary gastric cancer analysed by Tamura et al. found that LOH at the MCC locus occurred in all cases though other studies have found much lower incidences (88,106-108). Sanz-Ortega et al. subsequently reported the loss of MCC heterozygosity in intestinal metaplasia and in dysplastic lesions, in addition to gastric cancer (109). LOH at the MCC locus is frequently accompanied by LOH at the APC locus and both are found in differentiated and undifferentiated, early and advanced gastric carcinoma (106). Interestingly, a study by Rhyu et al. found that allelic deletion at MCC/APC was not detected in tumours that did not have allelic deletion of TP53, suggesting that MCC allelic deletions do not occur independently of TP53 alterations in gastric carcinogenesis (110).
FAT atypical cadherin 4 (FAT4)
FAT4 suppresses tumour growth via activation of Hippo signalling (111,112). In a recent study by Jung et al., loss of FAT4 protein expression was found in 24% of gastric cancer cases and was associated with increased invasiveness, high TNM stage and reduced disease-free survival time (111).
Zinc finger, MYND-type containing 10 (ZMYND10)
ZMYND10 is involved in the regulation of gastric cancer cell proliferation and colony formation (113). Methylation of ZMYND10 is significantly higher in gastric cancer compared with normal tissues with subsequent reduction in ZMYND10 protein expression levels (114).
Ubiquitin protein ligases
The CHFR gene codes for E3 ubiquitin-protein ligase and abnormal promoter methylation of this gene is thought to play a role in gastric carcinogenesis (115). The carboxyl terminus of Hsc-70-interacting protein (CHIP) is a U-box-type ubiquitin ligase, the overexpression of which is associated with reduced gastric cancer growth (116).
Controversial TSGs in gastric cancer
Runt-related transcription factor 3 (RUNX3), a member of the RUNX transcription factor family, is a downstream effector of the TGFβ signaling pathway which regulates cell proliferation, apoptosis, angiogenesis, adhesion and invasion (117). Several studies have found that loss of function of RUNX3 is associated with gastric carcinogenesis and this effect appears to be mediated via the Akt1, TGFβ and Wnt signaling pathways (7,117-119). Loss of RUNX3 is also significantly associated with the presence of H. pylori Cag (+), spontaneous EMT and is associated with the degree of gastric mucosal inflammation, atrophy and intestinal metaplasia (7,117). The expression of RUNX3 also correlates with that of E-cadherin (7).
Lu et al. found that the proportion of gastric lesions with RUNX3 promoter methylation increased with increasing stage of gastric cancer progression, with promoter methylation evident in 16% of chronic atrophic gastritis, 37% of intestinal metaplasia, 42% in gastric adenoma, 55% in dysplasia and 75% in gastric cancer (120). These effects were more pronounced in H. pylori positive patients compared to those who were H. pylori negative (120).
Despite these features, however, Lotem et al. have recently re-appraised the evidence regarding the role of RUNX3 and assert that it is not a bone fide cell-autonomous TSG although it may have an indirect influence on tumour development by virtue of its important roles in immunity and inflammation (121).
Epigenetic changes in gastric cancer
Altered gene expression can occur not only as a consequence of genetic or environmental factors but as a result of epigenetic factors resulting in gastric carcinogenesis (122,123). These inheritable, but non-genetic chromosomal modifications, which include DNA methylation and histone modification, occur as a result of chromatin modifiers and non-coding RNA (ncRNA) and result in altered gene expression (123). Inactivation of TSGs in gastric cancer is more commonly due to epigenetic change by promoter methylation than it is due to mutation (124).
For example, epigenetic inactivation of hepatocyte growth factor (HGF) activator inhibitor type 2 (HAI-2) is found in gastric cancer (125). Loss or reduction in expression of the iroquois homeobox 1 (IRX1) TSG on chromosome 5p15.33 is also found in gastric cancer and work by Guo et al. has shown that this is not due to mutations in IRX1, but rather, it is due to hypermethylation suppressing the transcription of IRX1 (126). Another study by Ling et al. found that N-myc downstream regulated gene 2 (Ndrg2), a candidate metastasis suppressor gene, is frequently downregulated in gastric cancer cell lines and tissues (127). Moreover, this effect, which occurs secondary to hypermethylation of the Ndrg2 promoter is associated with H. pylori infection and it is thought that H. pylori induces this effect by NF-κB activation and up-regulating DNMT3b (127).
Non-coding RNAs (ncRNAs)
ncRNAs regulate gene expression and are essential for the control of important cell functions including cell proliferation and survival (128). Deregulation of ncRNAs contributes to gastric cancer development by altering the expression of oncogenes or TSGs and by their regulatory effect on factors such as PTEN, E-cadherin, Akt and p53 (128,129). They have been shown to play a major role in gastric cancer development, invasion and metastasis (130). They can be sub-divided into long non-coding (lncRNAs) and miRNAs (128).
miRNAs are small, endogenous single stranded RNA molecules that negatively regulate gene expression by cleaving target mRNA or inhibiting translation (131). They play a significant role in the development of gastric cancer via the regulation of cell cycle progression, invasion, motility, metastasis and apoptotic cell death (129,132). Altered expression of miRNAs has been associated with gastric tumour size, degree of differentiation, disease stage and the presence of metastasis (130). Several previous studies have suggested that several miRNAs act as tumour suppressors in gastric carcinogenesis via a number of mechanisms including deregulation of E-cadherin (133-142).
lncRNAs are non-coding transcripts over 200 nucleotides long that have also been shown to act as oncogenes and tumour suppressors and may play an important role in the development of gastrointestinal carcinomas (143,144). They regulate gene expression by chromosome remodeling, transcriptional or post-transcriptional gene processing, resulting in a range of effects including imprinting, gene activation, gene repression and cell proliferation (145-147). In gastric cancer, lncRNAs have been implicated in the regulation of cell proliferation, cell-cell adhesion, EMT, extracellular matrix degradation, cell migration, invasion, metastasis, angiogenesis and tumour cell apoptosis (128,148,149).
Piwi-interacting RNAs (piRNAs) are a recently discovered class of small ncRNAs that bind specifically to Piwi protein family members to form piRNA compounds (piRC) that regulate gene-silencing pathways (130). They are implicated in gastric carcinogenesis and have shown promise as a possible peripheral blood-based diagnostic biomarker (150).
H. pylori and TSGs in gastric cancer
H. pylori promotes gastric carcinogenesis by a variety of means: it stimulates oxygen-derived free radical release from activated neutrophils, increases cytidine deaminase activity in infected gastric tissues resulting in mutations, CagA is secreted into gastric epithelial cells causing inflammation and oncogene activation, it induces epigenetic transformations such as promoter methylation in TSGs, and it induces aberrant expression of miRNAs (151).
Cumulative loss of TSGs in gastric cancer
There is a wealth of evidence to suggest that many TSGs may play a role in gastric cancer individually but also evidence to suggest that cumulative loss or abnormalities in these genes may also be important. A study by Cho et al. investigating simultaneous LOH in advanced gastric cancer, found that 33% of tumours informative at APC, DCC and RB1 loci had LOH at all three of these loci (89).
Gene therapy and TSG in gastric cancer
Improved understanding of the TSGs involved in gastric carcinogenesis has opened up potential opportunities for novel therapies involving the reintroduction of TSGs by gene therapy using viral or synthetic vectors (152). In particular, adenoviruses provide an attractive vehicle for gene therapy and have been used in phase III clinical trials to deliver wild-type TP53 to patients with head and neck and ovarian cancer (153). The reintroduction of TSGs by gene therapy is an attractive treatment strategy since it offers the prospect of cancer treatment without toxicity to nearby normal cells (154). However, there are currently a number of issues limiting the clinical application of gene therapy including concerns regarding toxicity, ethical and regulatory issues, practical limitations such as efficiency of gene transfer as well as limitations in disease response, which need to be overcome (155).
Reintroduction of TSGs by gene therapy aims to reinstate the cell’s natural regulatory mechanism for suppressing tumour growth but may have additional therapeutic effects, such as inducing apoptosis via the activation of other genes and increasing the sensitivity of cancer cells to chemotherapy (156,157).
Previous studies have shown that infection of human gastric carcinoma cell lines with recombinant TP53 adenovirus vector results in inhibition of growth and apoptotic cell death (157,158). Meanwhile, a review by Ishii et al. found that reintroduction of FHIT inhibited in vitro tumour cell growth or tumourigenicity in 57% of experiments analysed (159). Moreover, experimental work by Ishii et al. has shown that in FHIT-deficient mice, orally introduced viral vector-mediated FHIT was associated with reduced tumour development and reversal of established tumours by apoptosis (160). Reintroduction of CDKN2A using an expression vector resulted in reduced gastric cancer cell growth rate and enhanced sensitivity to chemotherapeutic drugs (161).
Conclusions
Gastric cancer is a complex, multifactorial disease in which TSGs play an essential role. Genetic and epigenetic changes in these genes contribute to gastric carcinogenesis regardless of histological type at all stages of disease progression and show promise as potential biomarkers of disease or as therapeutic targets.
Acknowledgements
The authors would like to acknowledge the support of our collaborative research partners in Peking University, Capital Medical University and Yiling Medical Research Institute.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- World Health Organization. All cancers (excluding non-melanoma skin cancer) estimated incidence, mortality and prevalence worldwide in 2012. Cited June 2015. Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx
- Gong J, Cao J, Liu G, et al. Function and mechanism of F-box proteins in gastric cancer (review). Int J Oncol 2015;47:43-50. [PubMed]
- Chun N, Ford JM. Genetic testing by cancer site: stomach. Cancer J 2012;18:355-63. [PubMed]
- Cancer research UK. Types of stomach cancer. Cited June 2015. Available online: http://www.cancerresearchuk.org/about-cancer/type/stomach-cancer/about/types-of-stomach-cancer
- Chiurillo MA. Role of the Wnt/β-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med 2015;5:84-102. [PubMed]
- Percesepe A, Ponz De Leon M. Hereditary factors in tumors of the digestive system. Ann Ist Super Sanita 1996;32:629-42. [PubMed]
- Wagih HM, El-Ageery SM, Alghaithy AA. A study of RUNX3, E-cadherin and β-catenin in CagA-positive Helicobacter pylori associated chronic gastritis in Saudi patients. Eur Rev Med Pharmacol Sci 2015;19:1416-29. [PubMed]
- Shiotani A, Cen P, Graham DY. Eradication of gastric cancer is now both possible and practical. Semin Cancer Biol 2013;23:492-501. [PubMed]
- Correa P, Shiao YH. Phenotypic and genotypic events in gastric carcinogenesis. Cancer Res 1994;54:1941s-1943s. [PubMed]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31-49. [PubMed]
- Solcia E, Fiocca R, Luinetti O, et al. Intestinal and diffuse gastric cancers arise in a different background of Helicobacter pylori gastritis through different gene involvement. Am J Surg Pathol 1996;20 Suppl 1:S8-22. [PubMed]
- Nakayama T, Ling ZQ, Mukaisho K, et al. Lineage analysis of early and advanced tubular adenocarcinomas of the stomach: continuous or discontinuous? BMC Cancer 2010;10:311. [PubMed]
- Kuzumaki N. Progress of research on tumor suppressor genes. Hokkaido Igaku Zasshi 1996;71:133-7. [PubMed]
- Cavenee WK, Dryja TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 1983;305:779-84. [PubMed]
- Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820-3. [PubMed]
- Cito L, Pentimalli F, Forte I, et al. Rb family proteins in gastric cancer (review). Oncol Rep 2010;24:1411-8. [PubMed]
- Bellacchio E, Paggi MG. Understanding the targeting of the RB family proteins by viral oncoproteins to defeat their oncogenic machinery. J Cell Physiol 2013;228:285-91. [PubMed]
- Poremba C, Bankfalvi A, Dockhorn-Dworniczak B. Tumor suppressor gene p53. Theoretical principles and their significance for pathology. Pathologe 1996;17:181-8. [PubMed]
- Triantafillou NG, Grosman IM, Verma RS. Genomania of p53 protein in gastric cancer. J Clin Gastroenterol 1996;22:170-3. [PubMed]
- NCBI. TP53 tumor protein p53 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/7157
- Xi Y, Chen Y. Oncogenic and Therapeutic Targeting of PTEN Loss in Bone Malignancies. J Cell Biochem 2015. [Epub ahead of print]. [PubMed]
- Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis 2004;9:667-76. [PubMed]
- NCBI. PTEN phosphatase and tensin homolog [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/5728
- NCBI. CDH1 cadherin 1, type 1 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/999
- NCBI. SMAD4 SMAD family member 4 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/4089
- NCBI. FHIT fragile histidine triad [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/2272
- Yousuf A, Bhat MY, Pandith AA, et al. MGMT gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell Oncol (Dordr) 2014;37:245-52. [PubMed]
- NCBI. MGMT O-6-methylguanine-DNA methyltransferase [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/4255
- NCBI. CD82 CD82 molecule [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/3732
- NCBI. CDKN2A cyclin-dependent kinase inhibitor 2A [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/1029
- NCBI. WWOX WW domain containing oxidoreductase [Homo sapiens (human)]. Cited June 2015. Avail
- Xing R, Li W, Cui J, et al. Gastrokine 1 induces senescence through p16/Rb pathway activation in gastric cancer cells. Gut 2012;61:43-52. [PubMed]
- NCBI. APC adenomatous polyposis coli [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/324
- NCBI. DCC DCC netrin 1 receptor [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/1630
- NCBI. RB1 retinoblastoma 1 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/592
- NCBI. PML promyelocytic leukemia [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/5371
- NCBI. KIAA1324 KIAA1324 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/57535
- NCBI. CEBPA CCAAT/enhancer binding protein (C/EBP), alpha [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/1050
- Lee HS, Lee HK, Kim HS, et al. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol 2003;200:39-46. [PubMed]
- Yokozaki H, Tahara E. Multistep stomach carcinogenesis. Nihon Rinsho 1996;54:1066-71. [PubMed]
- Shinozaki-Ushiku A, Kunita A, Fukayama M. Update on Epstein-Barr virus and gastric cancer (review). Int J Oncol 2015;46:1421-34. [PubMed]
- Yau TO, Tang CM, Yu J. Epigenetic dysregulation in Epstein-Barr virus-associated gastric carcinoma: disease and treatments. World J Gastroenterol 2014;20:6448-56. [PubMed]
- Yasui W, Nakayama H, Kuniyasu H, et al. New prognostic factors in human gastric carcinomas. Gan To Kagaku Ryoho 1992;19:26-33. [PubMed]
- Sano T, Tsujino T, Yoshida K, et al. Frequent loss of heterozygosity on chromosomes 1q, 5q, and 17p in human gastric carcinomas. Cancer Res 1991;51:2926-31. [PubMed]
- Saavedra K, Valbuena J, Olivares W, et al. Loss of Expression of Reprimo, a p53-induced Cell Cycle Arrest Gene, Correlates with Invasive Stage of Tumor Progression and p73 Expression in Gastric Cancer. PLoS One 2015;10:e0125834. [PubMed]
- Ochiai A, Yamauchi Y, Hirohashi S. p53 mutations in the non-neoplastic mucosa of the human stomach showing intestinal metaplasia. Int J Cancer 1996;69:28-33. [PubMed]
- Tamura G, Kihana T, Nomura K, et al. Detection of frequent p53 gene mutations in primary gastric cancer by cell sorting and polymerase chain reaction single-strand conformation polymorphism analysis. Cancer Res 1991;51:3056-8. [PubMed]
- Hamada M, Fujiwara T, Hizuta A, et al. The p53 gene is a potent determinant of chemosensitivity and radiosensitivity in gastric and colorectal cancers. J Cancer Res Clin Oncol 1996;122:360-5. [PubMed]
- André AR, Ferreira MV, Mota RM, et al. Gastric adenocarcinoma and Helicobacter pylori: correlation with p53 mutation and p27 immunoexpression. Cancer Epidemiol 2010;34:618-25. [PubMed]
- Pacchiani N, Censini S, Buti L, et al. Echoes of a distant past: The cag pathogenicity island of Helicobacter pylori. Cold Spring Harb Perspect Med 2013;3. [PubMed]
- Xu WT, Yang Z, Lu NH. Roles of PTEN (Phosphatase and Tensin Homolog) in gastric cancer development and progression. Asian Pac J Cancer Prev 2014;15:17-24. [PubMed]
- Wen YG, Wang Q, Zhou CZ, et al. Mutation analysis of tumor suppressor gene PTEN in patients with gastric carcinomas and its impact on PI3K/AKT pathway. Oncol Rep 2010;24:89-95. [PubMed]
- Carneiro P, Fernandes MS, Figueiredo J, et al. E-cadherin dysfunction in gastric cancer--cellular consequences, clinical applications and open questions. FEBS Lett 2012;586:2981-9. [PubMed]
- Corso G, Carvalho J, Marrelli D, et al. Somatic mutations and deletions of the E-cadherin gene predict poor survival of patients with gastric cancer. J Clin Oncol 2013;31:868-75. [PubMed]
- Zeng W, Zhu J, Shan L, et al. The clinicopathological significance of CDH1 in gastric cancer: a meta-analysis and systematic review. Drug Des Devel Ther 2015;9:2149-57. [PubMed]
- Yakirevich E, Resnick MB. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am 2013;42:261-84. [PubMed]
- Wei B, Shi H, Lu X, et al. Association between the expression of T-cadherin and vascular endothelial growth factor and the prognosis of patients with gastric cancer. Mol Med Rep 2015;12:2075-81. [PubMed]
- Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev 2000;11:49-58. [PubMed]
- Huebner K, Garrison PN, Barnes LD, et al. The role of the FHIT/FRA3B locus in cancer. Annu Rev Genet 1998;32:7-31. [PubMed]
- Kastury K, Baffa R, Druck T, et al. Potential gastrointestinal tumor suppressor locus at the 3p14.2 FRA3B site identified by homozygous deletions in tumor cell lines. Cancer Res 1996;56:978-83. [PubMed]
- Rocco A, Schandl L, Chen J, et al. Loss of FHIT protein expression correlates with disease progression and poor differentiation in gastric cancer. J Cancer Res Clin Oncol 2003;129:84-8. [PubMed]
- Imai Y, Oda H, Nakatsuru Y, et al. A polymorphism at codon 160 of human O6-methylguanine-DNA methyltransferase gene in young patients with adult type cancers and functional assay. Carcinogenesis 1995;16:2441-5. [PubMed]
- Zhao C, Bu X. Promoter methylation of tumor-related genes in gastric carcinogenesis. Histol Histopathol 2012;27:1271-82. [PubMed]
- Park TJ, Han SU, Cho YK, et al. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer 2001;92:2760-8. [PubMed]
- Hibi K, Sakata M, Yokomizo K, et al. Methylation of the MGMT gene is frequently detected in advanced gastric carcinoma. Anticancer Res 2009;29:5053-5. [PubMed]
- Zou XP, Zhang B, Zhang XQ, et al. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol 2009;40:1534-42. [PubMed]
- Lee JH, Seo YW, Park SR, et al. Expression of a splice variant of KAI1, a tumor metastasis suppressor gene, influences tumor invasion and progression. Cancer Res 2003;63:7247-55. [PubMed]
- Hinoda Y, Adachi Y, Takaoka A, et al. Decreased expression of the metastasis suppressor gene KAI1 in gastric cancer. Cancer Lett 1998;129:229-34. [PubMed]
- Tsutsumi S, Shimura T, Morinaga N, et al. Loss of KAI1 expression in gastric cancer. Hepatogastroenterology 2005;52:281-4. [PubMed]
- Tamura G. Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histol Histopathol 2004;19:221-8. [PubMed]
- Aldaz CM, Ferguson BW, Abba MC. WWOX at the crossroads of cancer, metabolic syndrome related traits and CNS pathologies. Biochim Biophys Acta 2014;1846:188-200.
- Maeda N, Semba S, Nakayama S, et al. Loss of WW domain-containing oxidoreductase expression in the progression and development of gastric carcinoma: clinical and histopathologic correlations. Virchows Arch 2010;457:423-32. [PubMed]
- Guo W, Dong Z, Dong Y, et al. Genetic and epigenetic alterations of WWOX in the development of gastric cardia adenocarcinoma. Environ Mol Mutagen 2013;54:112-23. [PubMed]
- Lewandowska U, Zelazowski M, Seta K, et al. WWOX, the tumour suppressor gene affected in multiple cancers. J Physiol Pharmacol 2009;60 Suppl 1:47-56. [PubMed]
- He D, Zhang YW, Zhang NN, et al. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr virus-associated gastric carcinomas. Med Oncol 2015;32:92. [PubMed]
- Yan J, Zhang M, Zhang J, et al. Helicobacter pylori infection promotes methylation of WWOX gene in human gastric cancer. Biochem Biophys Res Commun 2011;408:99-102. [PubMed]
- Oien KA, McGregor F, Butler S, et al. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J Pathol 2004;203:789-97. [PubMed]
- Yoon JH, Choi WS, Kim O, et al. The role of gastrokine 1 in gastric cancer. J Gastric Cancer 2014;14:147-55. [PubMed]
- Moss SF, Lee JW, Sabo E, et al. Decreased expression of gastrokine 1 and the trefoil factor interacting protein TFIZ1/GKN2 in gastric cancer: influence of tumor histology and relationship to prognosis. Clin Cancer Res 2008;14:4161-7. [PubMed]
- Yoon JH, Song JH, Zhang C, et al. Inactivation of the Gastrokine 1 gene in gastric adenomas and carcinomas. J Pathol 2011;223:618-25. [PubMed]
- Yoon JH, Kang YH, Choi YJ, et al. Gastrokine 1 functions as a tumor suppressor by inhibition of epithelial-mesenchymal transition in gastric cancers. J Cancer Res Clin Oncol 2011;137:1697-704. [PubMed]
- Mao W, Chen J, Peng TL, et al. Downregulation of gastrokine-1 in gastric cancer tissues and restoration of its expression induced gastric cancer cells to apoptosis. J Exp Clin Cancer Res 2012;31:49. [PubMed]
- Tamura G, Ogasawara S, Nishizuka S, et al. Two distinct regions of deletion on the long arm of chromosome 5 in differentiated adenocarcinomas of the stomach. Cancer Res 1996;56:612-5. [PubMed]
- Nagase H, Nakamura Y. Mutations of the APC (adenomatous polyposis coli) gene. Hum Mutat 1993;2:425-34. [PubMed]
- Tamura G. Molecular pathogenesis of adenoma and differentiated adenocarcinoma of the stomach. Pathol Int 1996;46:834-41. [PubMed]
- Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ 2004;327-49. [PubMed]
- Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol 2006;12:192-8. [PubMed]
- Liu B, Zhang X, Yu W. Relation between loss of heterozygosity at APC and MCC genetic loci and biological behaviour of gastric carcinoma. Zhonghua Zhong Liu Za Zhi 1996;18:3-5. [PubMed]
- Cho JH, Noguchi M, Ochiai A, et al. Loss of heterozygosity of multiple tumor suppressor genes in human gastric cancers by polymerase chain reaction. Lab Invest 1996;74:835-41. [PubMed]
- Tahara E. Molecular biology of gastric cancer. World J Surg 1995;19:484-8. [PubMed]
- Monden T, Yamamoto H, Ikeda K, et al. RB gene expression in gastrointestinal tract. Nihon Rinsho 1996;54:965-71. [PubMed]
- Li F, Liu J, Li S. MicorRNA 106b~25 cluster and gastric cancer. Surg Oncol 2013;22:e7-10. [PubMed]
- Xu Z, Chen J, Shao L, et al. Promyelocytic leukemia protein enhances apoptosis of gastric cancer cells through Yes-associated protein. Tumour Biol 2015. [Epub ahead of print] [PubMed]
- Kang JM, Park S, Kim SJ, et al. KIAA1324 suppresses gastric cancer progression by inhibiting the oncoprotein GRP78. Cancer Res 2015. [Epub ahead of print]. [PubMed]
- Regalo G, Resende C, Wen X, et al. C/EBP alpha expression is associated with homeostasis of the gastric epithelium and with gastric carcinogenesis. Lab Invest 2010;90:1132-9. [PubMed]
- NCBI. MCC mutated in colorectal cancers [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/4163
- NCBI. CPEB1 cytoplasmic polyadenylation element binding protein 1 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/64506
- NCBI. ADAMTS9 ADAM metallopeptidase with thrombospondin type 1 motif, 9 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/56999
- NCBI. PAX5 paired box 5 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/5079
- NCBI. ZFP82 ZFP82 zinc finger protein [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/284406
- NCBI. BCL6B B-cell CLL/lymphoma 6, member B [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/255877
- NCBI. FAT4 FAT atypical cadherin 4 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/79633
- NCBI. ZMYND10 zinc finger, MYND-type containing 10 [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/51364
- NCBI. CHFR checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/55743
- NCBI. STUB1 STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase [Homo sapiens (human)]. Cited June 2015. Available online: http://www.ncbi.nlm.nih.gov/gene/10273
- Tamura G, Maesawa C, Suzuki Y, et al. Primary gastric carcinoma cells frequently lose heterozygosity at the APC and MCC genetic loci. Jpn J Cancer Res 1993;84:1015-8. [PubMed]
- Hsieh LL, Huang YC. Loss of heterozygosity of APC/MCC gene in differentiated and undifferentiated gastric carcinomas in Taiwan. Cancer Lett 1995;96:169-74. [PubMed]
- Sud R, Talbot IC, Delhanty JD. Infrequent alterations of the APC and MCC genes in gastric cancers from British patients. Br J Cancer 1996;74:1104-8. [PubMed]
- Sanz-Ortega J, Sanz-Esponera J, Caldes T, et al. LOH at the APC/MCC gene (5Q21) in gastric cancer and preneoplastic lesions. Prognostic implications. Pathol Res Pract 1996;192:1206-10. [PubMed]
- Rhyu MG, Park WS, Jung YJ, et al. Allelic deletions of MCC/APC and p53 are frequent late events in human gastric carcinogenesis. Gastroenterology 1994;106:1584-8. [PubMed]
- Jung HY, Cho H, Oh MH, et al. Loss of FAT Atypical Cadherin 4 Expression Is Associated with High Pathologic T Stage in Radically Resected Gastric Cancer. J Gastric Cancer 2015;15:39-45. [PubMed]
- Katoh M. Function and cancer genomics of FAT family genes (review). Int J Oncol 2012;41:1913-8. [PubMed]
- Xiao K, Yu Z, Shi DT, et al. Inactivation of BLU is associated with methylation of Sp1-binding site of BLU promoter in gastric cancer. Int J Oncol 2015. [Epub ahead of print]. [PubMed]
- Shao Y, Zhang W, Zhang C, et al. High-resolution melting analysis of BLU methylation levels in gastric, colorectal, and pancreatic cancers. Cancer Invest 2010;28:642-8. [PubMed]
- Hu SL, Huang DB, Sun YB, et al. Pathobiologic implications of methylation and expression status of Runx3 and CHFR genes in gastric cancer. Med Oncol 2011;28:447-54. [PubMed]
- Liu F, Zhou J, Zhou P, et al. The ubiquitin ligase CHIP inactivates NF-κB signaling and impairs the ability of migration and invasion in gastric cancer cells. Int J Oncol 2015;46:2096-106. [PubMed]
- Subramaniam MM, Chan JY, Yeoh KG, et al. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta 2009;1796:315-31.
- Lin FC, Liu YP, Lai CH, et al. RUNX3-mediated transcriptional inhibition of Akt suppresses tumorigenesis of human gastric cancer cells. Oncogene 2012;31:4302-16. [PubMed]
- Voon DC, Wang H, Koo JK, et al. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells 2012;30:2088-99. [PubMed]
- Lu XX, Yu JL, Ying LS, et al. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer 2012;118:5507-17. [PubMed]
- Lotem J, Levanon D, Negreanu V, et al. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta 2015;1855:131-43.
- Necula LG, Mambet C, Albulescu R, et al. Epigenetics in gastric carcinogenesis: tet genes as important players. J Immunoassay Immunochem 2015;36:445-55. [PubMed]
- Kang C, Song JJ, Lee J, et al. Epigenetics: an emerging player in gastric cancer. World J Gastroenterol 2014;20:6433-47. [PubMed]
- Otani K, Li X, Arakawa T, et al. Epigenetic-mediated tumor suppressor genes as diagnostic or prognostic biomarkers in gastric cancer. Expert Rev Mol Diagn 2013;13:445-55. [PubMed]
- Dong W, Chen X, Xie J, et al. Epigenetic inactivation and tumor suppressor activity of HAI-2/SPINT2 in gastric cancer. Int J Cancer 2010;127:1526-34. [PubMed]
- Guo X, Liu W, Pan Y, et al. Homeobox gene IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene 2010;29:3908-20. [PubMed]
- Ling ZQ, Ge MH, Lu XX, et al. Ndrg2 promoter hypermethylation triggered by helicobacter pylori infection correlates with poor patients survival in human gastric carcinoma. Oncotarget 2015;6:8210-25. [PubMed]
- Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget 2014;5:8027-38. [PubMed]
- Wan X, Ding X, Chen S, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol 2015;36:521-32. [PubMed]
- Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol 2014;20:5411-9. [PubMed]
- Hua HB, Yan TT, Sun QM. miRNA polymorphisms and risk of gastric cancer in Asian population. World J Gastroenterol 2014;20:5700-7. [PubMed]
- Jiang C, Chen X, Alattar M, et al. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther 2015;22:291-301. [PubMed]
- Li X, Li H, Zhang R, et al. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem 2015;35:2033-42. [PubMed]
- Zuo QF, Zhang R, Li BS, et al. MicroRNA-141 inhibits tumor growth and metastasis in gastric cancer by directly targeting transcriptional co-activator with PDZ-binding motif, TAZ. Cell Death Dis 2015;6:e1623.
- Li Z, Yu X, Wang Y, et al. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget 2014. [Epub ahead of print]. [PubMed]
- Song P, Wang W, Tao G, et al. A miR-29c binding site genetic variant in the 3'-untranslated region of LAMTOR3 gene is associated with gastric cancer risk. Biomed Pharmacother 2015;69:70-5. [PubMed]
- Guo X, Guo L, Ji J, et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun 2010;398:1-6. [PubMed]
- Pan J, Hu H, Zhou Z, et al. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol Rep 2010;24:105-12. [PubMed]
- Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 2010;20:784-93. [PubMed]
- Gong Y, Ren J, Liu K, et al. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J Gastroenterol 2015;21:2949-58. [PubMed]
- Kang W, Tong JH, Lung RW, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer 2015;14:52. [PubMed]
- Carvalho J, van Grieken NC, Pereira PM, et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol 2012;228:31-44. [PubMed]
- Wang GY, Zhu YY, Zhang YQ. The functional role of long non-coding RNA in digestive system carcinomas. Bull Cancer 2014;101:E27-31. [PubMed]
- Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet 2014;5:164. [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [PubMed]
- Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337-46. [PubMed]
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159-66. [PubMed]
- Yang ZG, Gao L, Guo XB, et al. Roles of long non-coding RNAs in gastric cancer metastasis. World J Gastroenterol 2015;21:5220-30. [PubMed]
- Ayesh S, Matouk I, Schneider T, et al. Possible physiological role of H19 RNA. Mol Carcinog 2002;35:63-74. [PubMed]
- Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem 2011;44:1050-7. [PubMed]
- Nishizawa T, Suzuki H. Gastric Carcinogenesis and Underlying Molecular Mechanisms: Helicobacter pylori and Novel Targeted Therapy. Biomed Res Int 2015;2015:794378.
- Szlosarek PW, Dalgleish AG. Potential applications of gene therapy in the patient with cancer. Drugs Aging 2000;17:121-32. [PubMed]
- Anklesaria P. Gene therapy: a molecular approach to cancer treatment. Curr Opin Mol Ther 2000;2:426-32. [PubMed]
- Khalighinejad N, Hariri H, Behnamfar O, et al. Adenoviral gene therapy in gastric cancer: a review. World J Gastroenterol 2008;14:180-4. [PubMed]
- Pandha HS, Martin LA, Rigg AS, et al. Gene therapy: recent progress in the clinical oncology arena. Curr Opin Mol Ther 2000;2:362-75. [PubMed]
- Xiao B, Shi YQ, Zhao YQ, et al. Transduction of Fas gene or Bcl-2 antisense RNA sensitizes cultured drug resistant gastric cancer cells to chemotherapeutic drugs. World J Gastroenterol 1998;4:421-5. [PubMed]
- Tatebe S, Matsuura T, Endo K, et al. Adenovirus-mediated transfer of wild-type p53 gene results in apoptosis or growth arrest in human cultured gastric carcinoma cells. Int J Oncol 1999;15:229-35. [PubMed]
- Ohashi M, Kanai F, Ueno H, et al. Adenovirus mediated p53 tumour suppressor gene therapy for human gastric cancer cells in vitro and in vivo. Gut 1999;44:366-71. [PubMed]
- Ishii H, Dumon KR, Vecchione A, et al. Potential cancer therapy with the fragile histidine triad gene: review of the preclinical studies. JAMA 2001;286:2441-9. [PubMed]
- Ishii H, Zanesi N, Vecchione A, et al. Regression of upper gastric cancer in mice by FHIT gene delivery. FASEB J 2003;17:1768-70. [PubMed]
- Jeong YW, Kim KS, Oh JY, et al. Exogenous wild-type p16INK4A gene induces delayed cell proliferation and promotes chemosensitivity through decreased pRB and increased E2F-1 expressions. Int J Mol Med 2003;12:61-5. [PubMed]


