Research on microRNAs leads to new frontiers of clinical and translational relevance for gastric cancer management
The microRNAs (miRNAs) are a new field of ongoing cancer research, motivating the enthusiasms of scientists from all over the world. It is our opinion that the recent paper by Tang et al., published by the journal Clinical Cancer Research represents an excellent evidence of this paradigm (1). miRNAs are small non-coding RNAs controlling gene expression that were initially discovered in 1993 in the nematode Caenorhabditis elegans (2).
Since then, the miRNAs were the objective of an increasing number of investigations; the first evidence of their involvement in human cancers was provided in 2002 with the studies conducted by Croce et al., on the chronic lymphocytic leukemia (3). Henceforward, several authors investigated the miRNAs expression through microarray or PCR analyses in different cancerous tissues and cell lines. Indeed, over the last few years, the assessment of miRNAs has been characterized by the definition of their profile and their targets in serum and different normal and cancerous tissues.
In this field, Volinia and colleagues conducted a large genomic analysis investigating several gastrointestinal (GI) cancers (including stomach, pancreas and colon cancers) and documented the miRNAs’ profiling as cancer-specific. According to their results, GI cancers seemed to have a distinct miRNA’s signature comparing with non-GI cancers as e.g., lung or breast neoplasms (4).
In addition, the miRNAs’ signatures have been documented as tissue-specific as they could identify the specific cancer tissue from where they originated, and thus specifically classify those GI cancers derived e.g., from the stomach versus liver, esophagus, colon or pancreas (5). Furthermore, the aberrant miRNAs’ expression profile has been documented as correlated with the occurrence, the development, and the prognosis of GI cancers (4).
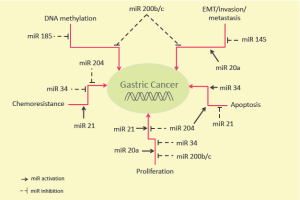
The study of miRNAs, however, usually involves the assessment of their molecular targets that are implicated in many important biological processes ranging from cell growth, to cell cycle, apoptosis, cell migration, senescence and chemoresistance (Figure 1). As miRNAs have multiple targets, their function in tumorigenesis could be either correlated to the regulation of a few or a number of specific targets and thus, specific pathways associated to cancer development (6).
Target-prediction algorithms can be used to identify the mRNA targets on the basis of (I) the complementarity between the mature miRNA’s sequence and the target; (II) the binding energy of the complex miRNA-target duplex; (III) the evolutionary conservation of the target site sequences and position in aligned UTRs of homologous genes (7).
However, this process has to be experimentally validated in order to eliminate the false positives, since the number of predicted sites is usually very large (6).
Intriguingly, also the identification of miRNAs’ targets involved in cancer and contributing to the malignant transformation could characterize different pathways that control miRNAs’ aberrant expression.
Nevertheless, if miRNA targets are crucial for the expression of the malignant phenotype and the cancer cells depend on target disregulation for proliferation and survival, we could expect that the use of miRNAs or anti-miRNAs molecules will lead to tumour regression (6).
In fact, as over the last few years we observed a shift from conventional chemotherapy to targeted therapies, we could speculate that miRNAs and anti-miRNAs will contribute in a short future to the development of advanced tailored therapies (6).
On the basis of this solid background, it seems clear that the characterization of miRNAs’ signature in the neoplastic tissues and in the serum samples of cancer patients might lead to new frontiers of high translational impact. This includes the early neoplastic detection, the clinical monitoring, the management of the prognosis, and possibly the development of new gene-based therapies or agents able to overcome the chemoresistance.
The identification of miRNAs’ signature associated with gastric cancer is nowadays a field of ongoing and intense research.
Gastric cancer is the 4th most common cancer worldwide (8), with more than 70% of cases occurring in the developing countries. Gastric carcinogenesis is a multistep process that involves many environmental and genetic factors, including the infection by Helicobacter pylori, as well as other genetic, dietary, chronic gastric inflammation-causing factors.
Recently, aberrant miRNAs’ expression has been associated with gastric carcinogenesis (9); indeed, several authors investigated the aberrant miRNA’s profiling (over-expression vs. down-regulation) in different types of gastric tumors and at different stages (Figure 1).
We recently profiled the expression of 851 human miRNAs in gastric tumours tissues and their matched peri-tumoural samples. This led to the identification of miRNA-204 as statistically and significantly down-regulated in cancerous tissues. According to our results, the down-regulation of miRNA204 was associated with the T stage of disease, since patients presenting a T1 stage displayed a lower down-regulation comparing to those with more advanced stage. On the basis of this background miRNA-204 might be used as a molecular biomarker for gastric cancer staging. The combination of the hysto-pathological (TNM) and molecular features (including e.g., miRNA204, Bcl-2, p53 status, ErbB2, c-myc) might strongly contribute to an accurate molecular profiling of gastric tumours (10).
As mentioned before, several mechanisms contribute to miRNAs’ aberrant expression during carcinogenesis, including genetic mutations, epigenetic silencing and a deregulated transcriptional activity.
It has been observed that several miRNAs could constitute a cluster of 2-7 genes controlled by the same regulatory sequences and whose expression might be highly similar (11); these different miRNAs could be considered altogether and investigated also as a family of interrelated non-coding RNAs.
One of the most investigated miRNA families is that of the miRNA200 which comprises five members (miRNA200a, miRNA200b, miRNA200c, miRNA141, and miRNA429) clustered and expressed as two separate polycistronic pre-miRNA transcripts; precisely the miRNA200b-200a-429 cluster is located at 1p36 and miRNA200c-141 cluster at chromosomal location 12p13.
Previous evidence has shown that the miRNA200 family is an important regulator of the epithelial-to-mesenchymal transition (EMT) system; the EMT has been described as a part of the embryonic development, but it has been also documented during the carcinogenesis process, when cancer cells shift from a differentiated to a more invasive and undifferentiated shape.
After EMT induction, cells loose the epithelial feature, acquire the flattened/mesenchymal characteristics (including vimentin filaments) and an invasive phenotype (by the expression of proteases which allow migration) and thus displaying all those crucial steps towards the metastatization process (12).
Because of their influence on the EMT process, the miRNA200 family has been recognized with a tumor suppressive role in a wide range of cancers, including breast (13), colorectal (14) pancreatic (14) and endometrial carcinomas (15); to date, however, its role in gastric cancer remains undefined.
The study presented from Tang and colleagues on “Clinical Cancer Research” analyzed the level of expression of miRNA200b and miRNA200c in 126 gastric cancer tissues and in adjacent normal gastric mucosae, as well as in eight gastric epithelial cell lines and in non-malignant gastric cells GES-1; authors detected that the expression of these miRNAs inversely correlated with the depth of invasion, the stage of the disease, and the presence of nodal metastases in gastric cancer patients; moreover the over-expression of either miRNA200b and miRNA200c markedly attenuated cell proliferation, migration ability and invasion of MGC-803 and AGS gastric cancer cells lines (1) (Figure 1). Furthermore, the down-regulation of miRNA200b and miRNA200c resulted as an independent predictor of worse overall and disease free survival for gastric cancer patients (1).
Notably, the serum concentration of miRNA200c has been documented as an epithelial-specific clinical biomarker useful for gastric cancer diagnosis and an independent prognostic marker for progression and survival of gastric cancer patients (16).
To understand the suppressive role of miRNA200b and miRNA200c in gastric cancer growth and invasion, Tang and colleagues used the TaregtScan and Miranda algorithms for putative mRNA targets. They identified that the DNA methyltransferases (DNMTs) DNMT3A and DNMT3B were predicted to be targets for miRNA200b and miRNA200c by both algorithms. DNA methyltransferases are enzymes involved in DNA methylation that cooperate in establishing and maintaining CpG-island methylation patterns; thereby playing a major role in the regulation of gene expression.
DNA methylation is a well-studied epigenetic phenomena, it plays a key role in X-chromosome inactivation, in the transcriptional silencing of foreign DNA elements and in the gene imprin
Methylation of CpG dinucleotide occurs in human cells when a methyl group is covalently added into the carbon-5' of CpG dinucleotide leading to the formation of 5' methylcytosine (5-mC); an aberrant DNA methylation pattern has been correlated to aging and chronic inflammation and it is implicated with viral infections and cancer development (17).
The methylation process is mediated by at least three active DNMTs: DNMT1 preferentially acts on hemimethylated CpG dinucleotide and it is necessary for the maintenance of specific methylation patterns during DNA replication, while DNMT3A and DNMT3B contribute to the methylation of unmodified DNA (18).
Tang and colleagues demonstrated that the transfection of miRNA200b or miRNA200c into MGC-803 and AGS cells markedly reduced the level of DNMT3A and DNMT3B proteins.
Furthermore, although DNMT1 was not a predicted target of miRNA200 family, the transfection of miRNA200b or miRNA200c into MGC-803 and AGS cells also reduced the level of the DNMT1 protein. The latter is frequently over-expressed in gastric cancers (19).
Further experiments documented that the down-regulation of DNMT1 was a consequence of the reduced activity of SP1, a zinc finger transcription factor that directly binds to the promoter of DNMT1 up-regulating the transcription (1).
Sp1 binds GC-rich elements that are common regulatory elements found in the promoters of several genes. Its expression is increased in a number of cancer cells including in those of gastric, breast and pancreatic carcinomas and it has been inversely correlated with the survival of gastric cancer patients (20).
According to the Targetscan 6.2 algorithm, the 3'UTR of Sp1 contains one predicted binding site for mRNA200b and miRNA200c through which miRNAs could down-regulate the protein expression and inhibit the DNMT1 transactivation.
Indeed, the restoration of miRNA200b and miRNA200c levels in MGC-803 and AGS cell lines resulted in a global DNA hypo-methylation that occurred through the direct binding of DNMT3A and DNMT3B to 3'UTR and only partially to an indirect effect on the DNMT1 promoter (1) (Figure 1).
The silencing of tumor suppressor genes by aberrant hyper-methylation is one of the earliest molecular events associated with cellular transformation that could be considered a predictor of tumor progression.
Many studies are evaluating the application of gene methylation status as a specific marker for allowing cancer diagnosis in biopsy specimens and non-invasive body fluids, such as serum or gastric washes.
The high prevalence of gene methylation, such as DAPK, CDH1, GSTP1, p15, and p16, has been documented in the serum of gastric cancer patients possibly due to the release of nucleic acid by gastric cancer cells, and it has been significantly correlated with the gene methylation in gastric cancer tissues.
Serum RASSF1A methylation has been documented significantly higher in gastric cancer patients comparing to those evidenced in benign gastric disease. The methylation of p16 promoter has been frequently detected in tumor samples, but not in matched normal tissues; moreover, p16 methylation is an early molecular event in gastric carcinogenesis. Therefore, the detection of methylated genes in serum may be a useful biomarker for early detection of gastric cancer (21).
According to this background, the DNA methylation would be an excellent target for anti-cancer therapies. It was found that accompanying DNA demethylation is a dramatic reactivation of the silenced genes and inhibition of cancer cell proliferation, promotion of cell apoptosis, or sensitization of cells to other chemotherapeutic reagents.
Several small natural and synthetic molecules are able to contrast the DNA hyper-methylation through inhibition of DNA methyl-transferase (DNMTi). Indeed, de-methylating agents are drugs which inhibit the methylation process and restore the expression of the previously hyper-methylated and silenced genes.
Emerging interest in the use of DNMTi as a potential strategy for cancer treatment is constantly increasing. Several small natural and synthetic molecules are widely used for in vitro studies and in clinical trials for their potential anti-cancer activities (22,23).
Cytidine analogs such as 5-azacytidine (azacitidine) and 5-azadeoxycytidine (decitabine) are the most commonly used demethylating agents. Both these drugs have been approved in the treatment of myelodysplastic syndrome (MDS) by Food and Drug Administration (FDA) in United States.
It seems important to highlight, however, that DNMTi for chemotherapy is still at a very early stage of progression, but nevertheless it is a field of ongoing researches and investigations. Those progress made in epigenetic research will lead to a better understanding of the actions of DNMTi, which will promote the translation from “bench to the bedside”. An ideal epigenetic therapy should be able to distinguish aberrantly methylated genes from normally methylated genes.
One of the most important findings reported by Tang and colleagues was that over-expression of miRNA200b and miRNA200c not only reduced gastric cancer cell proliferation and invasion but also global DNA methylation restoring the expression of p16, E-cadherin and RASSF1A (1).
Indeed, it is our opinion that the investigation of miRNAs also in gastric cancer tissues is moving forward to the identification of new frontiers of clinical use (Table 1): the ultimate comprehension of the tumorigenesis and of the network of genetic alterations involved in tumor’s development would contribute to tailor more personalized cancer treatments.
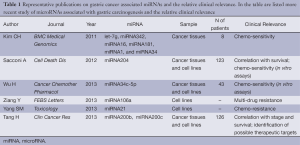
Full Table
Indeed as stated by a recent comprehensive review by Iorio (24) the potential of miRNAs' expression to correlate with the response to different therapies needs to be further investigated and validated by in vivo studies aiming to the definition of chemosensitivity or conversely to drug resistance. However, since the vast majority of the literature in this field reports preclinical studies, this area of investigation still represents an open question for future research. This process can be speed pursuing interdisciplinary cooperation between basic, translational and clinical scientists.
Acknowledgements
Financial support: Laura Lorenzon holds a Post-doctoral Fellowship Grant 2014 from Fondazione Veronesi; Valeria Canu holds a fellowship from AIRCS (Associazione Italiana Ricerca Colangite Sclerosante).
Disclosure: The authors declare no conflict of interest.
References
- Tang H, Deng M, Tang Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res 2013;19:5602-12. [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002;99:15524-9. [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006;103:2257-61. [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [PubMed]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704-14. [PubMed]
- John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol 2004;2:e363. [PubMed]
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137-50. [PubMed]
- Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology 2012;143:35-47.e2.
- Sacconi A, Biagioni F, Canu V, et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis 2012;3:e423. [PubMed]
- Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 2002;21:4663-70. [PubMed]
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894-907. [PubMed]
- Yu Y, Wu J, Guan L, et al. Kindlin 2 promotes breast cancer invasion via epigenetic silencing of the microRNA200 gene family. Int J Cancer 2013;133:1368-79. [PubMed]
- Paterson EL, Kazenwadel J, Bert AG, et al. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia 2013;15:180-91. [PubMed]
- Bai JX, Yan B, Zhao ZN, et al. Tamoxifen represses miR-200 microRNAs and promotes epithelial-to-mesenchymal transition by up-regulating c-Myc in endometrial carcinoma cell lines. Endocrinology 2013;154:635-45. [PubMed]
- Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med 2012;10:186. [PubMed]
- Wagner JR, Busche S, Ge B, et al. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol 2014;15:R37. [PubMed]
- Yang J, Wei X, Wu Q, et al. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol Med Rep 2011;4:1139-43. [PubMed]
- Etoh T, Kanai Y, Ushijima S, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol 2004;164:689-99. [PubMed]
- Deniaud E, Baguet J, Chalard R, et al. Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PLoS One 2009;4:e7035. [PubMed]
- Qu Y, Dang S, Hou P.. Gene methylation in gastric cancer. Clin Chim Acta 2013;424:53-65. [PubMed]
- Ren J, Singh BN, Huang Q, et al. DNA hypermethylation as a chemotherapy target. Cell Signal 2011;23:1082-93. [PubMed]
- Lewandowska J, Bartoszek A.. DNA methylation in cancer development, diagnosis and therapy--multiple opportunities for genotoxic agents to act as methylome disruptors or remediators. Mutagenesis 2011;26:475-87. [PubMed]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4:143-59. [PubMed]


