The six-ankyrin repeat containing Asb genes may drive normal and adenomatous compartment size expansion in the distal intestine
Abstract
Dysregulation of compartment size is maybe the most fundamental biological issue in cancer. Important clues as to novel therapeutical avenues for dealing with this disease may thus be obtained from the regulation of compartment size in untranstormed cells. Previously we reported the extraction of d-asb11 from a screen in zebrafish to identify gene products essential for maintaining proliferation progenitors during embryogenesis and established that it is a major determinant regulating the size of the central nervous system; knock down of the gene leading to a small brain, whereas its forced expression led to a massive expansion of the zebrafish central nervous system and brain hypertrophy. Here we compare zebrafish d-asb11 to the human genome and identify a subfamily within the group of Asb genes consisting apart from human ASB11 of ASB5, ASB9, and ASB13 which are highly structurally related, very homologous and among the most conserved genes in the chordate phylum. Analysis of forced expression of d-asb11 in zebrafish suggests the gene has the capacity to expand endodermal domains and thus we explore the expression of Asb5, Asb9, Asb11 and Asb13 in the untransformed mammalian gastrointestinal system and possible deregulation in gastrointestinal cancer. We observe low but distinct expression of various Asbs in the different elements of the gastrointestinal tract suggesting Asb-coding within the endoderm may exist. Asb9 appears most important, especially in the distal tract and its expression is highly upregulated in colonic cancer. The study indicates that Asb11-like members of the Asb family of proteins might be implicated in compartment expansion in the gastrointestinal tract and prompt further investigations as to their roles in cancer in derivatives of the endodermal lineage.
Key words
Compartment expansion; Asb genes; gastrointestinal tract; cancer
Introduction
Regulation of compartment size is one of the most fundamental issues in biology and especially cancer medicine (1,2). During embryogenesis cells become committed to specific cell fates, which are subsequently followed by expansion phases in which progenitor cells divide (3). When final compartment size is reached, cells exit the cell cycle and further differentiation occurs. Differences in compartment size represent an important driving force in evolution and in taxonomic descriptions of species, whereas the understanding of the forces that regulate compartment size defines one of the large outstanding questions in science. Furthermore, aberrant compartment regulation is implicated in many serious pathologies (most dramatically, maybe, in cancer) and thus knowledge as to the molecular factors that determine and maintain proliferation in progenitors is hardly a pursuit only of academic interest. Hence understanding the molecular mechanisms governing compartment size in the vertebrate body is a highly relevant subject. Nevertheless the constituting components acting in this process remain largely obscure.
In an effort to discover the genetic participants involved in compartment size regulation, we performed a screen in Danio rerio (zebrafish) to identify gene products whose presence is essential for maintaining proliferation of neural progenitors during embryogenesis (4). To this end zebrafish embryos were either left untreated or pulsetreated with 0.5 M all-trans retinoic acid (RA) between 50% and 60% epiboly when neural induction and patterning are on the way. Subsequently mRNA was isolated from wild type and RA-treated embryos at 90 % epiboly, which following conversion into cDNA was used for a differential display employing the transcripts from the untreated embryos as driver cDNA and the transcripts from the RA-treated embryos as tester cDNA. The resulting differential fragments were tested using whole mount in situ hybridization at different developmental stages from this screen and one fragment was singled out for detailed characterization based on its restricted spatiotemporal expression pattern during late gastrulation and early somitogenesis. Obtaining full-length sequence of the down-regulated 0.34 kb fragment revealed that the gene was a homologous to mammalian ankyrin and SOCS-box containing protein 11 (asb11) (2,5). Morpholino-mediated loss-of-function of by knock down of d-ASB11 in zebrafish abrogated the expansion phase in the central nervous system and induced premature neuronal differentiation followed by a reduced size of the definitive neuronal compartment and a small brain. Conversely, misexpression of d-asb-11 mRNA diminished or entirely abolished terminally differentiated primary neurons, and increased the size of the proneural compartment, producing prominent brain hypertrophy. Overexpression in PC12 cells abrogated the NGF-induced block of cell proliferation and inhibited nerve growth factor (NGF)-induced neurite extension, however d-Asb-11 did not interfere with NGF-induced commitment to the neuronal lineage. We concluded that Asb-11 is an important novel regulator of the size of the neuronal progenitor cell compartment (see Figure 1A).
Cues as to the possible molecular action of Asb-11 come from the presence of the conserved SOCS box motif and ankyrin repeats (6-9). The suppressors of cytokine signaling (SOCS) box is a structural C-terminal domain that interacts with Elongin C by its B/C box conserved motif. In turn, Elongin C binds Elongin B forming a dimer that bridges the substrate bound by the SOCS box protein to a Cullin protein. In turn, Elongin C binds Elongin B forming a dimer that bridges the substrate bound by the SOCS box protein to a Cullin protein. This step is thought to be supported by a conserved Cullin box motif, located downstream of the B/C box in the SOCS box. Cullin then recruits a RING-finger-containing protein Rbx1/2, completing the assembly of the ECS E3-type ubiquitin ligase complex. Therefore SOCS box proteins are the substrate recognition units of the ECS complexes, regulating the turnover of a wide range of proteins by targeting them for polyubiquitination and, subsequent, proteasomal degradation (10,11). SOCS box proteins are classified into different families based on the protein motifs found N-terminally of the SOCS box. Accordingly, we generated a zebrafish mutant line lacking the Cul5 box of the Ankyrin repeat and SOCS-box containing protein 11 (ASB11Cul) allowing us to explore the biological functions of the cullin box domain of d-ASB11 (12). Animals homozygous for this allele are defective for proper cell fate specification within the neurogenic regions of zebrafish embryos. To our knowledge, this is the first report of a metazoan mutant presenting a mutation in the consensus sequence of any SOCS box containing protein. Important mechanical insight as to how Asb proteins may effect compartment size comes from experiments in which we addressed the role of d-asb11 in Notch signalling. In canonical Delta/Notch signalling, expression of Delta activates Notch in neighbouring cells leading to down regulation of Delta in these neighbouring cells (13). This process of lateral inhibition results in selection of either Delta signalling cells or Notch signalling cells. In zebrafish embryos morpholino-mediated knockdown of d-ASB11 results in repression of different Delta/Notch elements and their transcriptional targets while these are induced upon d-ASB11 misexpression. d-ASB11 activates Notch reporters cell non-autonomously in vitro and in vivo when co-expressed with a Notch reporter, but represses Notch reporters when expressed in the DeltaA expressing cell. In agreement, d-ASB11 was able to specifically ubiquitinate and degrade DeltaA both in vitro and in vivo. Thus we concluded that d-Asb11 is a novel component of canonical Delta/Notch signalling (14).
Importantly, good evidence exists that Notch signalling may drive compartment size in gastrointestinal cancer as well (15,16), although in general it is fair to say that we do not understand the molecular drivers for aberent compartment size in gastrointestinal cancer. The mechanisms that maintain stemness in cancers are now well-understood and involve usually mutations in Wnt signalling cascade (17), but there is little evidence that Wnt signalling drives the compartment expansion that follows aberrant stemness. Mutations in p21Ras or associated signalling undoubtly play an important role here, but the relative lack of success of monotherapy with inhibitors of this signalling suggests we miss important players here and prompt identification of other candidate pathways here (18). Intriguingly, recently it was reported that the level of Asb9 expression correlated with patient survival in colorectal cancer (18,19), prompting furthers study whether Asb9 might constitute a mammalian d-Asb11 homologue and whether such homologues may have functionality in the gastrointestinal system. Here we report a first attempt to establish whether human d-Asb11 homologues might constitute such a pathway.
Methods
Zebra fish microinjection and whole mount in situ hybridisation
Zebrafish were kept at 27.5 ℃. Embryos were obtained by natural matings, cultured in embryo medium and staged according to Kimmel (20). Synthetic capped cRNAs of asb-11 or of control pT7TS globin-GFP construct were synthesized using the mMESSAGE mMACHINE kit (Ambion) according to the manufacturers’ instructions. Each of the cRNAs was injected as 250 pg in 0.5 nL into one-cell stage embryos. After 48 hrs fish were sacrificied and whole mount in situ hybridizations were performed according to Joore (21). Western Blots were performed according to Versteeg et al. (22,23).
RNA isolation and qRT–PCR
Total RNA extraction and purification was performed by using standard Trizol and isopropanol precipitation. cDNA synthesis was performed using hexamer primers and MLV Reverse Transcriptase. Transcript levels were quantified by real-time PCR using ABsolute QPCR SYBR Green Fluorescein Mix (Westburg) on an iCycler iQ Real-Time PCR detection system (Bio-Rad) as described earlier (24). Results are expressed as a relative ratio to the housekeeping gene actin and represent a representive image of at least three independent experiments, each consisting of at least 8 animals. Asb11-F: GTCAGAAGGCCTGGACCAT and Asb11-R: CTCATGGAGTGGGGATCG. Asb9-F: TCCTCTTCATGATGCTGCAA and Asb9-R: CACGTGATCTGCTGTGATGA.
Bioinformatical analysis
Sequences of asb homologues were downloaded from NCBI and relative homologies of either the entire sequence or the domain N-terminal from the first ankyrin repeat were determined using clustalw (25) to determine relative distances of each sequence. Subsequently, visualisation of these distances as provided in Figure 1 was performed in Coreldraw. For analysis of asb transcript numbers in human disease the GDS2947 dataset as provided through GEO was employed, giving transcriptomes of 32 adenomas and 32 normal mucosa samples from the same patients (reference). In addition the GDS3121 and GD1321 data depositories were used, also as provided by GEO. These sets provide an expression profiling atlas of human and murine tissues and were thus employed to investigate expression patterns of different six ankyrin repeat containing Asb genes in the mammalian gastrointestinal tract.
Results
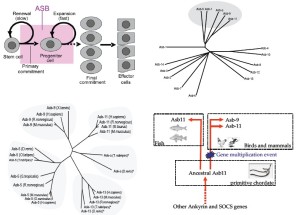
Definition of a six ankyrin-containing subset of Asb genes
The Asb family represents a chordate-restricted (no representatives in other phyla) but highly conserved family of intracellular proteins. Recently we identified zebrafish Asb11 (d-Asb11) as a potential regulator of compartment size of the central nervous system and the musculature. The high sequence homology of the ASB family of proteins, especially within the ankyrin repeats, makes it difficult to perform meaningful phylogenetic analysis on ASB family members, but it is recognised that the N-terminal half of ASB proteins shows greater general conservation than the C-teminal (26). Thus we performed a phylogentic analysis on the C-terminus of the ASB family to identify the position of d-Asb11 within this group of proteins. The C terminus of ASB-a clustered together with mammalian ASB-5, -9, -11 and -13, suggesting that d-Asb11 together with ASB-5, 9, 11 and 13 represents a distinctive subgroup within the Asb family of proteins (Figure 1B). In apparent agreement, the primary sequence of d-Asb11, ASB-a, -5, -9, -11 and -13 all contain 6 ankyrin repeats, setting these ASB proteins apart from the other ASB proteins which contain other numbers of ankyrin repeats. Searches in predicted amino acid sequences suggest a homologue of d-Asb11 in the teleost T. rubripes (Figure 1C). Thus d-Asb11 together with its mammalian homologues ASB-5, -9, -11 and -13 appear to form a distinct subfamily within the ASB family of proteins, probably sharing a common genetic precursor in an ancestral vertebrate, but with as yet unidentified functions in cellular physiology. Also analysis of the intron/ exon boundaries strongly supports the notion that these six ankyrin repeat domain containing proteins are a separate subgroup within the ASB family of proteins (5). In mammals and birds the two most close orthologues of d-asb11 are Asb9 and Asb11 which are highly homologous, both sharing approximately 70% sequence similarity to d-asb11. Asb9 and Asb11 lie adjacent to each other on the X-chromosome suggesting a sarcopterygiic-specific gene duplication event, not present in teleosts (Figure 1D). Furthermore Asb5, Asb9 and Asb11, but not Asb1, Asb2, or Asb15, activate a Notch reporter construct in Hela cells demonstrating that members of the six ankyrin repeat subfamily of Asb proteins share functional characteristics that at least certain other Asb family members not share with these six ankyrin repeat domain containing ASB proteins. Interestingly the six ankyrin repeat domain-containing proteins are the only the members of the ASB family of proteins of which representatives are known in non-vertebrate chordates and in fact Asb5, Asb9, Asb11 and Asb13 are among the most conserved genes in the chordate phylum, Ciona intestinalis and human Asb11 sharing approximately 49% homology in the coding sequence and even higher in the 3’ UTR, suggesting a fundamental importance of these genes in chordate physiology and we initiated experiments to define possible functionality of this family for compartment size in the gastrointestinal tract.
Overexpression of d-Asb11 drives compartment expansion in ectodermal lineages in the developing zebrafish
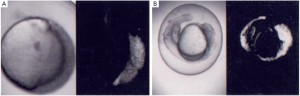
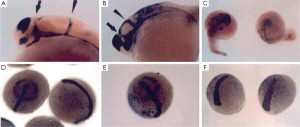
Evidence that Asb11 homologues may be involved in compartment size in the gastrointestinal tract came for experiments that examined the effects of forced expression of d-Asb11 by injecting mRNA in zebrafish zygotes. Even distribution of injected mRNAs was obtained by investigating the distribution of green fluorescent protein (GFP) rnRNA during development (Figure 2). Subsequently, expression patterns of marker genes in wildtype embryos as compared to embryos injected with d-Asb11 mRNA was compared. Figure 3 shows the effects of such forced d-Asb11 expression on of shh, emx-1 and wnt-1 and laxial, emx-1 and wnt-1 triple stained embryos. Especially Shh, axial and Wnt-I are well suited to stain the developing gastrointestinal system.. From the stainings it seems evident that not only the central nervous system but also the presumptive gastrointestinal system has expanded because of forced d-Asb11 expression. Although a crude form of experimentation, it thus seems that at least in the zebrafish d-asb11 can guide compartment size decisions in the gastrointestinal system and experimentation was initiated to investigate the expression of its homologues in higher vertebrates.
6-ankyrin repeat contain Asb gene-coding within the vertebrate endoderm may exist
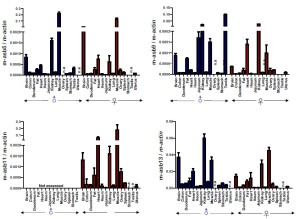
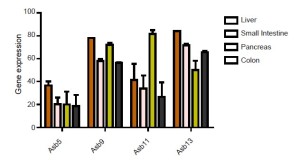
We investigated specific-tissue expression of murine Asb5, Asb9, Asb11 and Asb13 in a variety of tissues. The results are depicted in Figure 4. Expression levels vary widely, suggesting that a tissue-specific Asb coding exists. In general, however, the Asb genes tested are not expressed in an important way in the intestine, with the exception of Asb9 which is highly expressed in distal intestinal structures. Analysis of intestinal expression of six ankyrin repeat containing Asb expression in human primary tissue is broadly consistent with the expression data in mice (Figure 5). As mammalian Asb9 and Asb11 are both manifestations of the unduplicated d-asb11 in the zebrafish, it is tempting to speculate that expanded gastrointestinal domain seen in zebrafish by forced expression of d-asb11 represents a functionality that is conserved in asb-9 in higher vertebrates.
Specific overexpression of Asb9 in human colonic adenoma
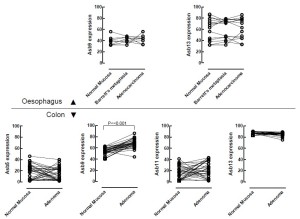
We downloaded the GDS3113 dataset from the Gene Expression Omnibus resource of the NCBI (http:// www.ncbi.nlm.nih.gov/geo/). This dataset contains the transcriptomes of 32 prospectively collected adenomas with those of normal mucosa from the same individuals. The transcriptomes were analysed for expression of ASB5,9,11, and -13 (Figure 6). It appears that normal colonic mucosa is characterised by low expression of ASB5 and ASB11, expression of ASB13 is higher, whereas ASB9 has a more intermediate expression level, broadly consistent with the results described above. When expression is compared between normal mucosa and the adjacent adenoma, expression of ASB5 and ASB11 does not show consistent alterations, expression of ASB13 appears somewhat decreased, however, strong upregulation of ASB9 observed. We also investigated ASB9 in oesophagal cancer, but no evidence for a role in cancer in this structure was obtained. Thus ASB9 is a strong candidate to drive compartment expansion in distal intestinal structures and cancer therein.
Discussion
In adult tissues, the size of the stem cell compartment is under tight genetic control (27). The stem cells, a restricted subpopulation of cells, continue proliferating and undergo either to self-renewal or differentiate into committed cell types with less proliferative potential. Deregulation of these processes result in uncontrolled cell growth and is implicated in many serious pathologies, as cancer (28). Hence to comprehend the biology of cancer, it is necessary to characterize the mechanisms that multicellular organisms evolved to control their proliferative capacity into appropriate compartments, and understand how these mechanisms are regulated in normal cells and how they become deregulated in cancer. Efforts to discover the participants involved in regulating compartment size have revealed gene products whose presence are essential for maintaining proliferation of progenitors during embryogenesis and adult life. One of these genes, which emerged from a genetic screen, was asb11 (Ankyrin repeat and SOCS box-containing protein 11). Asb11 was downregulated at the start of terminal cell commitment in zebrafish embryos and subsequent experiments revealed that Asb11 is a major determinant regulating the size of the central nervous system (5). As the asb11 gene, together with the highly homologous asb5, asb9, and asb13 are among the most conserved genes in the chordate phylum (Figure 1), we and others hypothesise that ASBs have a vertebrate subphylum-wide significance for maintaining expansion of progenitor compartments. In the present study we provide evidence that Asb coding of intestinal derivatives exists, that six ankyrin repeat containing Asb genes can drive compartment expansion in the endoderm and that in colon cancer Asb9 expression is upregulated. Expression of ASB9 in colorectal cancer has been reported before and shown in a multivariate analysis to be an independent prognostic factor of overall survival (relative risk, 4.09; 95% confidence interval, 1.47-11.88; P=0.007), but how these results relate to the present results remains unresolved (19). Thus we propose that studies as to the role of Asb9 in driving increased compartment size in colorectal cancer are warranted.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Giglio P, Gilbert MR. Neurologic complications of cancer and its treatment. Curr Oncol Rep 2010;12:50-9.
- Tee JM, Peppelenbosch MP. Anchoring skeletal muscle development and disease: the role of ankyrin repeat domain containing proteins in muscle physiology. Crit Rev Biochem Mol Biol 2010;45:318-30.
- Marks PA, Rifkind RA, Gambari R, et al. Commitment to terminal differentiation and the cell cycle. Curr Top Cell Regul 1982;21:189-203.
- Bink RJ, Habuchi H, Lele Z, et al. Heparan sulfate 6-o-sulfotransferase is essential for muscle development in zebrafish. J Biol Chem 2003;278:31118-27.
- Diks SH, Bink RJ, van de Water S, et al. The novel gene asb11: a regulator of the size of the neural progenitor compartment. J Cell Biol 2006;174:581-92.
- Kile BT, Viney EM, Willson TA, et al. Cloning and characterization of the genes encoding the ankyrin repeat and SOCS box-containing proteins Asb-1, Asb-2, Asb-3 and Asb-4. Gene 2000;258:31-41.
- Linossi EM, Nicholson SE. The SOCS box-Adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life 2012;64:316-23.
- Nicholson SE, Hilton DJ. The SOCS proteins: a new family of negative regulators of signal transduction. J Leukoc Biol 1998;63:665-8.
- Yoshida K. Identification and characterization of ankyrin repeat and SOCS box-containing gene ASB15 in silico. Int J Mol Med 2005;16:343-7.
- Piessevaux J, Lavens D, Peelman F, et al. The many faces of the SOCS box. Cytokine Growth Factor Rev 2008;19:371-81.
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009;78:399-434.
- Sartori da Silva MA, Tee JM, Paridaen J, et al. Essential role for the d-Asb11 cul5 Box domain for proper notch signaling and neural cell fate decisions in vivo. PLoS One 2010;5:e14023.
- Chitnis AB. The role of Notch in lateral inhibition and cell fate specification. Mol Cell Neurosci 1995;6:311-21.
- Diks SH, Sartori da Silva MA, Hillebrands JL, et al. d-Asb11 is an essential mediator of canonical Delta-Notch signalling. Nat Cell Biol 2008;10:1190-8.
- Miyamoto S, Rosenberg DW. Role of Notch signaling in colon homeostasis and carcinogenesis. Cancer Sci 2011;102:1938-42.
- Vooijs M, Liu Z, Kopan R. Notch: architect, landscaper, and guardian of the intestine. Gastroenterology 2011;141:448-59.
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011;17:313-9.
- Crul M, de Klerk GJ, Swart M, et al. Phase I clinical and pharmacologic study of chronic oral administration of the farnesyl protein transferase inhibitor R115777 in advanced cancer. J Clin Oncol 2002;20:2726-35.
- Tokuoka M, Miyoshi N, Hitora T, et al. Clinical significance of ASB9 in human colorectal cancer. Int J Oncol 2010;37:1105-11.
- Kimmel CB, Ballard WW, Kimmel SR, et al. Stages of embryonic development of the zebrafish. Dev Dyn 1995;203:253-310.
- Joore J, van der Lans GB, Lanser PH, et al. Effects of Retinoic Acid on the Expression of Retinoic Acid Receptors During Zebrafish Embryogenesis. Mechanisms of Development 1994;46:137-50.
- Braat H, Stokkers P, Hommes T, et al. Consequence of functional Nod2 and Tlr4 mutations on gene transcription in Crohn’s disease patients. J Mol Med (Berl) 2005;83:601-9.
- Versteeg HH, Sorensen BB, Slofstra SH, et al. VIIa/tissue factor interaction results in a tissue factor cytoplasmic domain-independent activation of protein synthesis, p70, and p90 S6 kinase phosphorylation. J Biol Chem 2002;277:27065-72.
- Muncan V, Heijmans J, Krasinski SD, et al. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat Commun 2011;2:452.
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002;Chapter 2:Unit 2.3.
- Kile BT, Viney EM, Willson TA, et al. Cloning and characterization of the genes encoding the ankyrin repeat and SOCS box-containing proteins Asb-1, Asb-2, Asb-3 and Asb-4. Gene 2000;258:31-41.
- Muller-Sieburg CE, Cho RH, Sieburg HB, et al. Genetic control of hematopoietic stem cell frequency in mice is mostly cell autonomous. Blood 2000;95:2446-8.
- Muller-Sieburg CE, Sieburg HB. Clonal diversity of the stem cell compartment. Curr Opin Hematol 2006;13:243-8.


