Effects of TUBB3, TS and ERCC1 mRNA expressions on chemoresponse and clinical outcome of advanced gastric cancer by multiplex branched-DNA liquid chip technology
Globally, gastric cancer (GC) is the fourth most commonly cancer and the second leading cause of cancer-related death (1). The advanced GC, which refers to the unresectable/recurrent GC, includes locally advanced unresectable GC (accounting for 30% of all GC), GC with distant metastasis (accounting for 30% of all GC), and recurrence after radical surgery (the recurrence rate ranges 40-60%). Therefore, up to 80% of GC patients will ultimately develop an advanced disease (1).
Currently there is still no standard first-line chemotherapy for the advanced GC. As shown in the V325 phase III trial, the docetaxel (TXT), cisplatin (CDDP), and fluorouracil (FU) (DCF) regimen had a response rate of 35-40% and extended the overall survival (OS) to 9 months (2). However, the limited survival gain, relatively high incidences of grade 3/4 toxicities, and individual sensitivities to the drugs during the treatment with the DCF regimen make the evidence-based standard chemotherapy, which is based on the results of population studies, difficult to be applied in clinical practice (3). Further studies on tumor molecular biology and pharmacogenomics have demonstrated that the expression levels of some specific genes in tumor tissues may influence the expressions and biological activities of some relevant proteins and thus regulate the metabolism of anticancer drugs and their action targets, which results in the individual sensitivities to the chemotherapy.
In our current study, by using multiplex branched-DNA liquid chip technology (4,5), we determined the mRNA expressions of β-tubulin-III (TUBB3), thymidylate synthase (TS), and excision repair cross complementation group 1 (ERCC1), which represent the resistance gene of FU, platinum-based antitumor drug, and paclitaxel, in the GC tissue, with an attempt to explore the influences of the expressions of these three genes on the treatment effectiveness, disease progression, and survival. We hope this study will help the clinicians to correctly identify patients who can really benefit from DCF regimen and predict the treatment outcomes. Also, this study may provide evidences for efficacy prediction, prognosis evaluation, and individualized treatment.
Subjects and methods
Subjects
Patients with advanced GC who were treated in the Department of Oncology, Xiangya Hospital, Central South University between June 2008 and June 2011 were screened. Patient who met the following inclusion criteria were enrolled in this retrospective study: (I) pathologically diagnosed as with gastric adenocarcinoma; (II) had distant metastases at diagnosis or recurrence after radical surgery; (III) tissue samples from primary tumors were collected before the initiation of chemotherapy; (IV) at least one measurable and evaluable target lesion was available; (V) having received at least two cycles of DCF as first-line palliative therapy (TXT 75 mg/m2, intravenous injection on day 1; CDDP 75 mg/m2, intravenous injection on day 1; and FU 750 mg/(m2.d), delivered as a continuous intravenous infusion using an ambulatory pump; and (VI) with complete clinicopathological and follow-up (till December 31, 2012) data.
Clinical data
Based on the inclusion criteria, a total of 48 pathologically confirmed advanced GC patients were enrolled, among whom there were 23 men and 25 women aged 26-79 years (median: 50 years). All of them had adenocarcinoma [15 with intestinal type and 33 with diffuse type, based on the Lauren classification (6)]. The measurable lesions were scanned using computed tomography (CT) or magnetic resonance imaging (MRI). No damage to vital organs was detected before treatment.
Methods
Detection of target genes
Using the multiplex branched-DNA liquid chip technology (Guangzhou Yishen Medical Inspection Institute), we detected the expressions of TUBB3, TS, and ERCC1 genes in the GC tissues. The paraffin-embedded tissues were cleaved to allow the release of RNA. Spherical particles in the kit were further used to capture the target RNA. After the RNA signal was amplified, Luminex® assay was applied to determine the expressions of the target mRNA. All the raw data obtained from the Luminex 200TM flow cytometry system were adjusted and normalized. The yielded netMFI values were the relative mRNA expressions of TUBB3, TS, and ERCC1.
Interpretation of the mRNA expressions of TUBB3, TS, and ERCC1
Using the median mRNA expression levels of TUBB3, TS, and ERCC1 as the thresholds, we divided the results as “high expression group” and “low expression group”.
Response evaluation
According to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, the treatment outcomes are divided as complete remission (CR), partial remission (PR), stable disease (SD) and progressive disease (PD). Patients with CR and PR were regarded as responders, and the overall remission rate (ORR) was calculated. Patients with PD were regard as non-responders. The evaluation of survival was based on time to progression (TTP, which was calculated from the stable status to the radiologically confirmed progression) and OS. Patients were also followed up via telephone.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 software. The associations of the clinicopathological features and the mRNA expression levels of TUBB3, TS, and ERCC1 with the chemotherapy response were analyzed with chi square test or Fisher’s exact test. The survival curves were drawn by using Kaplan-Meier method. The associations of the clinicopathological features and the mRNA expression levels of TUBB3, TS, and ERCC1 with TTP/OS were analyzed with Log-rank method. Cox proportional hazards models were applied for the multivariate analysis of the prognostic factors. A 2-sided P value less than 0.05 was considered statistically significant (α=0.05).
Results
mRNA expressions of TUBB3, TS, and ERCC1
The medians of the netMFI values of TUBB3, TS, and ERCC1 were 0.12499 (0.00765-1.63016), 0.11818 (0.00001-0.66684), and 0.89932 (0.31266-2.18099), respectively. Based on these medians, the patients were further divided as “high expression group” and “low expression group”.
Chemotherapy response
Relationship between clinicopathological features and chemotherapy response
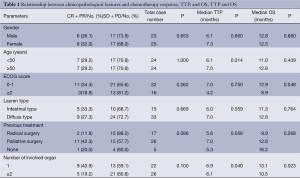
In our cohort, the ORR was 29.2% (14/48), where CR 2.1% (n), PR 27.1% (n=13), SD 58.3% (n=28), and PD 12.5% (n=6). The patients’ gender, age, performance status (ECOG) score, the Lauren type of tumors, and the number of involved organs showed no significant impacts on the chemotherapy response (Table 1).
Full Table
Relationships between the mRNA expression levels of TUBB3, TS, and ERCC1 and the chemotherapy response
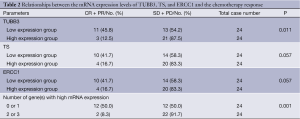
The patients with low TUBB3 mRNA expression had significantly higher response rate to chemotherapy than patients with high TUBB3 expression (45.8% vs. 12.5%, P=0.011). Patients with low TS and ERCC1 mRNA expressions also showed higher response rates, although not statistically significant (both P=0.057). Pooled analysis of the expression levels of these three genes showed that: patients with 0 or 1 gene high expression had a response of up to 50%, while those with high expressions of 2 or 3 genes had a response rate of only 8.3% (P=0.001) (Table 2).
Full Table
Prognosis
Relationship between clinicopathological features and TTP/OS
No patient was lost to follow-up. At the end of follow-up, all the 48 patients experienced disease progression, and 42 of them died. The median TTP of this cohort was 6.9 months (95% CI: 5.8-8.0 months) and the median OS was 12.5 months (95% CI: 10.5-14.5 months). The median OS was 8.9 months in patients with a ECOG score of ≥2 and 12.9 months in those with an ECOG score of 0 or 1 (P=0.048). The median TTP and median OS was 6.1 and 10.5 months, respectively, in patients with two or more organs being involved, and was 6.9 and 13.1 months, respectively, in patients with only one organ being involved (TTP: P=0.040; OS: P=0.023). The patients’ gender and age, the Lauren type of tumors, and the previous treatment showed no impact on TTP and OS (Table 1).
Relationships between the mRNA expression levels of TUBB3, TS, and ERCC1 and TTP/OS
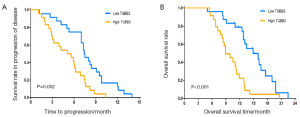
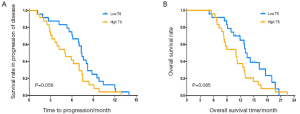
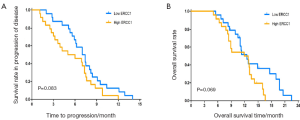
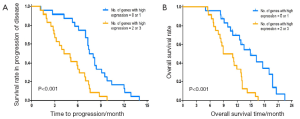
The median TTP and median OS were 7.4 and 14.9 months in patients with low mRNA express ion of TUBB3, which were significantly superior to those in patients with high mRNA expression of TUBB3 (5.1 and 8.9 months, respectively (TTP: P=0.002; OS: P<0.001) (Figure 1). The mRNA expression level of TS showed no significant impact on TTP and OS (TTP: P=0.056; OS: P=0.085) (Figure 2). Similarly, the mRNA expression level of ERCC1 was not significantly correlated with TTP and OS (TTP: P=0.083; OS: P=0.069) (Figure 3). The median TTP and median OS was 7.5 and 14.9 months, respectively, in patients with 0 or 1 organ being involved, and was 4.2 and 8.9 months, respectively, in patients with 2 or 3 organ being involved (TTP: P=0.001; OS: P=4) (Figure 4).
Prognostic factors
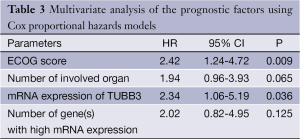
The prognostic factors including ECOG score, number of involved organs, mRNA expression level of TUBB3, and number of highly expressed TUBB3/TS/ERCC1 genes that had been found to be correlated with OS in the univariate analysis were further analyzed in the Cox proportional hazards regression model, which showed that an ECOG score of ≥2 (P=0.009) and high TUBB3 expression (P=0.036) were the independent adverse prognostic factors of OS, with the hazard ratio (HR) being 2.42 and 2.34, respectively (Table 3).
Full Table
Discussion
The multiplex branched-DNA liquid chip technology provides a novel approach for the quantitative detection of gene expression levels. It is highly sensitive, easy to perform, and also featured by wide linear range and high throughput. Therefore, it is feasible for the determination of various samples including cell, tissue, whole blood, blood stains, and paraffin-embedded sections. When used together with the LuminexTM systems, this technology can simultaneously determine multiple genes in a simple and flexible way. Compared with the conventional methods, the liquid chip technology has many advantages. For instance, it can directly detect the sample lysis solution without RNA retraction and reverse Transcription; also, no amplification of the target fragment is required. Therefore, the liquid chip technology can remarkably decrease the impacts of accumulated errors during multiple procedures or steps on the detection results. This technology uses a group of specific probes to capture mRNA segments and then amplify the signals, avoiding the application of polymerase chain reaction (PCR). Thus, even when the mRNA becomes broken down, the relevant information can still be detected. It can quantitatively detect 100 target genes, among which a group of reference genes can be included. It can be expected that the branched-DNA liquid chip technology will gradually replace the florescence quantitative PCR and exerts an increasingly important role in the determination of mRNA expressions of specific genes in tumor tissues to facilitate the tailored treatment. Using this technology, Ren et al. (7) evaluated the EGFR, K-RAS, and B-RAF somatic mutations and the mRNA expression of EGFR in the tumor paraffin-embedded specimens obtained from 14 patients with non-small cell lung cancer (NSCLC) who were receiving icotinib hydrochloride treatment in a phase I clinical trial as predicators of the efficacy of icotinib hydrochloride. They found that this technology was reliable for measuring EGFR mutation with high throughput and rapidity; meanwhile, the EGFR mutation status that was rapidly and effectively detected by the technology was a predictive biomarker for response to icotinib hydrochloride. Zhou et al. (8) applied this novel multiplex branched-DNA liquid chip technology to detect the mRNA expression levels of ERCC1, BRCA1, TUBB3, and STMN1 in 24 patients who were receiving platinum-based chemotherapy and 19 patients who were receiving platinum-based chemotherapy plus antimicrotubule chemotherapy. They found that patients with high ERCC1 and BRCA1 mRNA expression levels tended to have poor response to platinum-based chemotherapy, and meanwhile the resistance to antimicrotubule chemotherapy was associated with the high TUBB3 and STMN1 mRNA expression levels.
TUBB3 is the only β-tubulin isotype that can make the microtubules become unstable. Many clinical trials have demonstrated that its abnormal expression is closely associated with the resistance to antimicrotubule chemotherapeutic agents (9,10). In their in vitro experiment, Kavallaris et al. (11,12) found that the decrease of TUBB3 increased the sensitivity to anti-microtubule drugs. Kamath et al. (13) also found that the overexpression of TUBB3 could lower the inhibitory effect of paclitaxel on the kinetics of microtubules and thus induced drug resistance. However, few studies have explored the role of TUBB3 in GC. Only one study that had enrolled 20 advanced GC patients who were receiving TXT-based chemotherapy (14) showed that the response rate was significantly lower in patients positive for TUBB3 expression than in the negative group; its association with TTP and OS was not reported. In our current study, we applied the novel branched-DNA liquid chip technology to detect the expressions of TUBB3, TS, and ERCC1 genes simultaneously. A total of 48 advanced GC patients undergoing DCF regimen were enrolled. The results showed that the response rate was significantly lower in patients with high TUBB3 mRNA expression than those with low TUBB3 mRNA expression, and the median TTP and OS were significantly superior in patients with low TUBB3 mRNA expression than those with high TUBB3 mRNA expression. In addition, Cox multivariate analysis showed high TUBB3 expression is an independent adverse prognostic factor in patients with advanced GC.
After its entry into cells, FU is converted to 5-fluoro-2'-deoxyuridylate (FdUMP), and the latter can be bound with TS and CH2THF, forming stable triple complexes, which can inhibit the activity of TS and prevent the binding of dUMP and thymidine phosphorylase (TP) and thus hampers the DNA synthesis (15). TS is the main target enzyme via which FU exerts its cytotoxic effect. In FU-containing chemotherapy regimens, the expression level of TS may be correlated with the response to treatment. According to the expert consensus reached in the 2009 World Gastrointestinal Cancer Conference, the TS mRNA expression level is helpful for predicting the response to FU in patients with colorectal cancer. However, the role of TS for predicting the chemotherapy efficacy and prognosis in advanced GC patients remains controversial. Using immunohistochemical techniques, Yeh et al. (16) detected the TS expression in advanced GC patients undergoing FU-containing chemotherapy and found that patients with low TS expression had significantly better response rate and OS than those with high TS expression. Our group had also performed a meta-analysis enrolling 15 studies (844 patients with advanced GC) and found that high TS expression was an adverse prognostic factor for OS in advanced GC patients undergoing FU-based chemotherapy (HR =1.43, 95% CI: 1.08-1.90); However, its association with the response rate was statistically significant (17). Similarly, our current study showed that the TS mRNA expression was not significantly associated with the response rate, TTP, and OS in advanced GC patients receiving DCF regimen. We believed that the sensitivity and resistance to FU may be jointly affected by multiple key enzymes including TS, TP, and orotate trausphosphoribosylase (OPRT) during the metabolism of FU, and the expression of a single gene may not be able to determine the efficacy of FU. Secondly, patients in our current cohort received the combined therapy, and some patients who were resistant to FU might have benefited from TXT and/or CDPP. For patients who cannot tolerate the combined chemotherapy and receive FU alone, the TS expression may serve as a predictor of chemotherapy outcomes and prognosis.
ERCC1 is an important gene involved in nucleotide excision repair (NER) of damaged DNA. Its expression products can be bound with the DNA repair enzyme-deficient complementary gene F to form a tight heterodimer. The latter is a specific endonuclease that connect with the 5' end and has the dual capabilities of identifying injuries and cleaving the 5' end; By doing so, it regulates (particularly, limits the speed of) the NER (18). Some previous studies have found that the ERCC1 expression level was negatively correlated with the sensitivity to platinum-based therapies and the prognosis. Using the immunohistochemical techniques, Olaussen et al. (19) explored the ERCC1 expression in 1,867 NSCLC patients who had received radical surgeries and found that CDDP-based adjuvant chemotherapy could significantly extend the OS of patients with negative ERCDC1 expression but could not improve the prognosis of patients with positive results. However, in the studies conducted by Jeong et al. (20) and Yun et al. (21), ERCC1 expression was not significantly correlated with either the response to platinum-based chemotherapy or the prognosis. In our current study, although the correlations of ERCC1 expression with the response to DCF regimen, TTP, and OS were not statistically significant, patients with low ERCC1 expression did showed certain advantages over those with high ERCC1 expression in terms of response rate and prognosis. Nevertheless, the role of ERCC1 in predicting the outcomes of platinum-containing palliative chemotherapy for advanced GC requires further studies with larger sample sizes.
In summary, our current study, for the first time, analyzed the combined predictive role of TUBB3, TS, and ERCC1 for chemotherapy in advanced GC patients. The results showed that patients with 0 or 1 gene high expression tended to have longer TTP and OS. The combined detection of these three genes will facilitate the clinicians to correctly identify advanced GC patients who can really benefit from DCF regimen and obtain a better prognosis. TUBB3 may be a good predictor of the prognosis of advanced GC patients undergoing DCF regimen. Since patients with high TUBB3 expression may be not responsive to anti-microtubule agents (e.g., paclitaxel), other first-line chemotherapy drugs such as FU, CDDP, and EPI may be applied to improve the treatment efficacy and prognosis and meanwhile minimize chemotherapy-related toxicities. However, our study was limited by its retrospective design and small sample size. Prospective randomized clinical trials with larger sample sizes are warranted to further explore the predictive role of TUBB3 in advanced GC.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [PubMed]
- Van Cutsem E, Moiseyenko VM, Tjulandin S, et al. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: a report of the V325 Study Group. J Clin Oncol 2006;24:4991-7. [PubMed]
- Lu Q, Luduena RF. Removal of beta III isotype enhances taxol induced microtubule assembly. Cell Struct Funct 1993;18:173-82. [PubMed]
- He F, Li Y, Lin Z. The detection about expression of ERCC1, TYMS, TUBB3, RRM1 and mutation rate of EGFR (E19/20) in non-small cell lung cancer patients. Chinese Journal of Clinicians 2011;5:1337-450.
- Zhang L, Yang H, Xu J. Gene expression significance in personalized medicine of non-small-cell lung cancer and gene expression analyzing platforms. Curr Drug Metab 2011;12:455-9. [PubMed]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31-49. [PubMed]
- Ren GJ, Zhao YY, Zhu YJ, et al. Tumor gene mutations and messenger RNA expression: correlation with clinical response to icotinib hydrochloride in non-small cell lung cancer. Chin Med J (Engl) 2011;124:19-25. [PubMed]
- Zhou Q, He J, Yang X, et al. RNA expression profiling of ERCC1, BRCA1, TUBB3, and STMN1 in Non-Small Cell Lung Cancer (NSCLC) by a multiplex branched DNA liquidchip technology (MBL) for predicting the efficacy of chemotherapy. J Clin Oncol 2010;28:abstr e21020.
- Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-16. [PubMed]
- Burkhart CA, Kavallaris M, Band Horwitz S. The role of beta-tubulin isotypes in resistance to antimitotic drugs. Biochim Biophys Acta 2001;1471:O1-9. [PubMed]
- Kavallaris M, Kuo DY, Burkhart CA, et al. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J Clin Invest 1997;100:1282-93. [PubMed]
- Kavallaris M, Burkhart CA, Horwitz SB. Antisense oligonucleotides to class III beta-tubulin sensitize drug-resistant cells to Taxol. Br J Cancer 1999;80:1020-5. [PubMed]
- Kamath K, Wilson L, Cabral F, et al. BetaIII-tubulin induces paclitaxel resistance in association with reduced effects on microtubule dynamic instability. J Biol Chem 2005;280:12902-7. [PubMed]
- Urano N, Fujiwara Y, Doki Y, et al. Clinical significance of class III beta-tubulin expression and its predictive value for resistance to docetaxel-based chemotherapy in gastric cancer. Int J Oncol 2006;28:375-81. [PubMed]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003;3:330-8. [PubMed]
- Yeh KH, Shun CT, Chen CL, et al. High expression of thymidylate synthase is associated with the drug resistance of gastric carcinoma to high dose 5-fluorouracil-based systemic chemotherapy. Cancer 1998;82:1626-31. [PubMed]
- Hu HB, Kuang L, Zeng XM, et al. Predictive value of thymidylate synthase expression in gastric cancer: a systematic review with meta-analysis. Asian Pac J Cancer Prev 2012;13:261-7. [PubMed]
- Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 2007;33:9-23. [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [PubMed]
- Jeong SH, Han JH, Kim JH, et al. Bax predicts outcome in gastric cancer patients treated with 5-fluorouracil, leucovorin, and oxaliplatin palliative chemotherapy. Dig Dis Sci 2011;56:131-8. [PubMed]
- Yun J, Kim KM, Kim ST, et al. Predictive value of the ERCC1 expression for treatment response and survival in advanced gastric cancer patients receiving cisplatin-based first-line chemotherapy. Cancer Res Treat 2010;42:101-6. [PubMed]